Back
Introduction: Prediction of response to treatment is important in clinical decision-making. However this is challenging because the LUTS population is heterogeneous, and factors associated with treatment response are poorly understood. We used novel artificial intelligence (AI) models to analyze the high-dimensional, sparse and mixed-type data collected on men and women with LUTS enrolled in the Symptoms of Lower Urinary Tract Dysfunction Research Network (LURN), for a deeper understanding of predictors of treatment response.
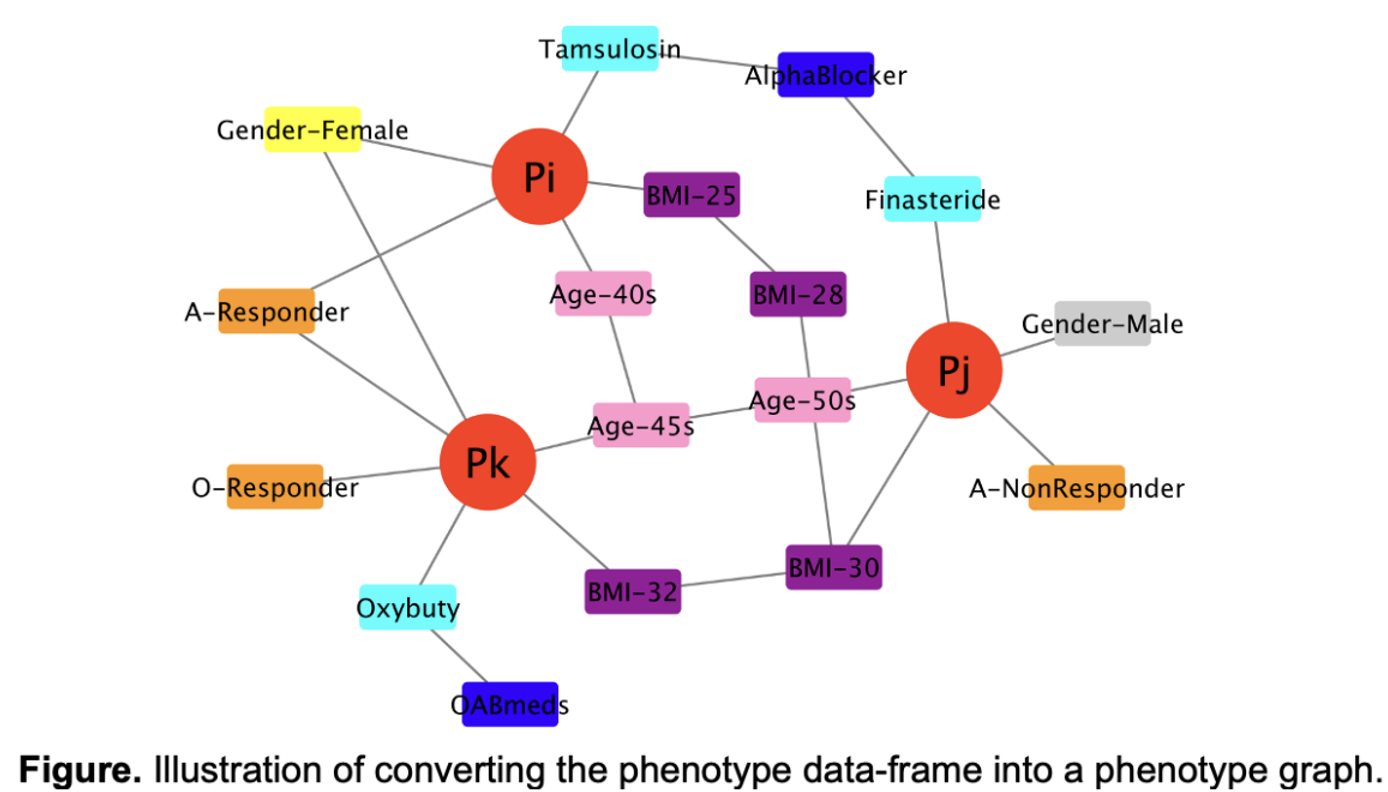
Methods: We developed a novel deep graph neural network (GNN) model LUTSPheNet to solve these challenges. First, the complex data were converted into a patient-phenotype graph that linked study participants with 181 phenotypic features. The “DeepWalk” model was used to convert the patient and phenotype nodes into phenotypically semantic numerical vectors (see Figure). The heterogenous phenotypic nodes with different data types were converted into the same numerical vector type, and were used as the input of an interpretable graph attention network (GAT) model to predict drug response of individual participants. Response to OAB medications (anticholinergics and/or beta-agonists) was classified as responders vs. non-responders based on changes in urgency urinary incontinence, urgency, or frequency symptom scores on the LUTS Tool.
Results: OAB medications were taken by 243 participants. Symptom data before and after treatment were available from 134. 87 (65%) were responders. Using the 5-fold cross validation, LUTSPheNet achieved a 71.6% accuracy in predicting responders. The top-ranked participant characteristics predicting response to OAB medications include: current smoker; incontinence during sleep; stress incontinence with physical activities; pain or discomfort in the bladder area; higher PROMIS anxiety, depression, constipation, diarrhea scores; and good pelvic floor contraction strength on Oxford Scale (females only).
Conclusions: Deep learning and artificial intelligence models are novel ways to identify clinical features predicting responders to OAB medications. A web-based prediction tool is being developed to help clinicians predict the probabilities of response to medications based on patient characteristics. SOURCE OF
Funding: NIH/NIDDK
Podium Session
Session: PD14: Urodynamics/Lower Urinary Tract Dysfunction/Female Pelvic Medicine: Female Incontinence: Therapy I
PD14-03: PREDICTION OF RESPONSE TO OAB MEDICATIONS USING DEEP LEARNING AND ARTIFICIAL INTELLIGENCE – A LURN RESEARCH STUDY
Saturday, April 29, 2023
7:20 AM – 7:30 AM CST
Location: S501A

Henry Henry Lai, MD
Professor of Urology and Anesthesiology
Washington University School of Medicine
Podium Presenter(s)
Introduction: Prediction of response to treatment is important in clinical decision-making. However this is challenging because the LUTS population is heterogeneous, and factors associated with treatment response are poorly understood. We used novel artificial intelligence (AI) models to analyze the high-dimensional, sparse and mixed-type data collected on men and women with LUTS enrolled in the Symptoms of Lower Urinary Tract Dysfunction Research Network (LURN), for a deeper understanding of predictors of treatment response.
Methods: We developed a novel deep graph neural network (GNN) model LUTSPheNet to solve these challenges. First, the complex data were converted into a patient-phenotype graph that linked study participants with 181 phenotypic features. The “DeepWalk” model was used to convert the patient and phenotype nodes into phenotypically semantic numerical vectors (see Figure). The heterogenous phenotypic nodes with different data types were converted into the same numerical vector type, and were used as the input of an interpretable graph attention network (GAT) model to predict drug response of individual participants. Response to OAB medications (anticholinergics and/or beta-agonists) was classified as responders vs. non-responders based on changes in urgency urinary incontinence, urgency, or frequency symptom scores on the LUTS Tool.
Results: OAB medications were taken by 243 participants. Symptom data before and after treatment were available from 134. 87 (65%) were responders. Using the 5-fold cross validation, LUTSPheNet achieved a 71.6% accuracy in predicting responders. The top-ranked participant characteristics predicting response to OAB medications include: current smoker; incontinence during sleep; stress incontinence with physical activities; pain or discomfort in the bladder area; higher PROMIS anxiety, depression, constipation, diarrhea scores; and good pelvic floor contraction strength on Oxford Scale (females only).
Conclusions: Deep learning and artificial intelligence models are novel ways to identify clinical features predicting responders to OAB medications. A web-based prediction tool is being developed to help clinicians predict the probabilities of response to medications based on patient characteristics. SOURCE OF
Funding: NIH/NIDDK

