Back
Purpose: In humans, there are 48 nuclear hormone receptors (NRs) that act as ligand-activated transcription factors to regulate various biological processes, such as metabolism, development, cell proliferation, immune response, central nervous system function, and reproduction. In addition to ligand binding, NR activation is highly controlled through co-regulatory proteins, in which coactivators increase transcription, and corepressors decrease target transcription. The Cummins laboratory discovered a new coactivator protein, arginine and glutamate rich 1 (ARGLU1)1 that significantly increases the activity of the metabolically important peroxisome proliferator-activated receptors (PPARs) (Figure 1)2. PPAR isoforms PPARα, PPARδ and PPARγ, are key modulators of lipid and carbohydrate metabolism and cellular differentiation. Therefore, we will examine the role of ARGLU1 in PPARα-regulated pathways including the fasting/refeeding cycle and under PPARα-specific ligand activation.
Methods: Since mouse knockouts of ARGLU1 were previously shown to be embryonically lethal, our laboratory produced floxed-Arglu1 mice (Arglu1fl/fl) and cross-bred these mice with Albumin-Cre mice to generate liver-specific Arglu1-null mice (Arglu1LKO).1 To determine if ARGLU1 could play a role in PPARα regulated pathways, we performed several in vivo experiments with Arglu1fl/fl and Arglu1LKO mice. When examining fasting and refeeding regulation, mice were housed in Promethion Metabolic Cages (Sable Systems) which measured food and water intake, physical activity, O2 consumption and CO2 production. This allowed the examination on whether changes in fuel utilization and energy expenditure are dependent on the hepatic expression of ARGLU1. The chow feeding conditions were either ad libitum fed, fasted for 24-hours, or fasted for 24 hours and refed for 24-hours. Plasma was collected for metabolite quantification. Hepatic gene expression was also examined for genes important to fasting and refeeding processes. When investigating the importance of liver Arglu1 upon PPARα activation, we treated 14-week-old Arglu1fl/fl and Arglu1LKO mice with 40 mg/kg WY14643 (a PPARα agonist) intraperitoneally for a 14-day period. Quadricep skeletal muscle, liver, intestine and epididymal white adipose tissues were flash frozen at -80°C for gene expression analysis. Liver RNA was isolated and target gene regulation (i.e., Cyp4a14, Ehhadh, Cd36, Fsp27, Fabp2, Pdk4) was examined via RT-qPCR. Plasma metabolites examined include glucose, cholesterol, triglycerides, glycerol, and non-esterified fatty acids.
Results: Results from the fasting and refeeding study demonstrate that ARGLU1 may have an important role in regulating the PPARα response to refeeding. Plasma glucose and triglyceride levels were unchanged between Arglu1fl/fl and Arglu1LKO mice (Figure 2a). Hepatic gene expression demonstrated the expected patterns of gene induction or repression upon fasting; however, there were no changes between Arglu1fl/fl and Arglu1LKO genotypes (Figure 2b). The lack of difference between genotypes in genes responsible for feeding regulation was surprising, as changes in the magnitude of PPARα target gene expression were anticipated based on previous studies performed in primary hepatocytes (Figure 1b). However, there was a significant difference between Arglu1fl/fl and Arglu1LKO refed groups in the gene regulation of Srebp-1c, a lipogenic regulator, which is highly increased upon refeeding, and dampened without ARGLU1 (Figure 2b). The respiratory exchange ratio (RER) followed the expected patterns of 0.9 upon feeding, indicating preferential glucose utilization, and around 0.7 RER with fasting when fat utilization is maximal for Arglu1fl/fl and Arglu1LKO groups (Figure 2c). Liver RNA was isolated from Arglu1fl/fl and Arglu1LKO mice treated with either vehicle or WY14643, and target gene regulation was examined via RT-qPCR. Specific genes involved in fatty acid utilization (Fgf21, Pdk4, Fabp2, Cd36) were dampened in the Arglu1LKO group upon WY14643 treatment (Figure 3a). However, not all genes were decreased, and some important fatty acid metabolic genes (Cyp4a14, Fsp27) were increased in Arglu1LKO mice (Figure 3b). No difference in plasma glucose levels was observed between genotypes (Figure 3c).
Conclusion: We conclude that ARGLU1 impacts select PPAR-regulated lipid metabolic pathways in unanticipated ways. The fasting response was not dramatically impacted; however, mice expressing ARGLU1 showed a tendency towards enhanced carbohydrate utilization on refeeding and warrants further investigation into its role in intermediary metabolism in vivo. Future directions also include examining the role of ARGLU1 in PPAR response to a high-fat diet.
References: 1. Magomedova L, Tiefenbach J, Zilberman E, Le Billan F, Voisin V, Saikali M, Boivin V, Robitaille M, Gueroussov S, Irimia M, Ray D, Patel R, Xu C, Jeyasuria P, Bader GD, Hughes TR, Morris QD, Scott MS, Krause H, Angers S, Blencowe BJ, Cummins CL. ARGLU1 is a transcriptional coactivator and splicing regulator important for stress hormone signaling and development. Nucleic Acids Res. 2019 Apr 8;47(6):2856-2870.
2. Cash SB., Sahin C, & Cummins CL. (2021). The role of liver arginine and glutamate rich 1 in coactivating PPAR nuclear receptor-mediated gene expression. American Association of Pharmaceutical Sciences PharmSci 360. Virtual.
Acknowledgments: We gratefully acknowledge funding for this project from the Canadian Institutes of Health Research.
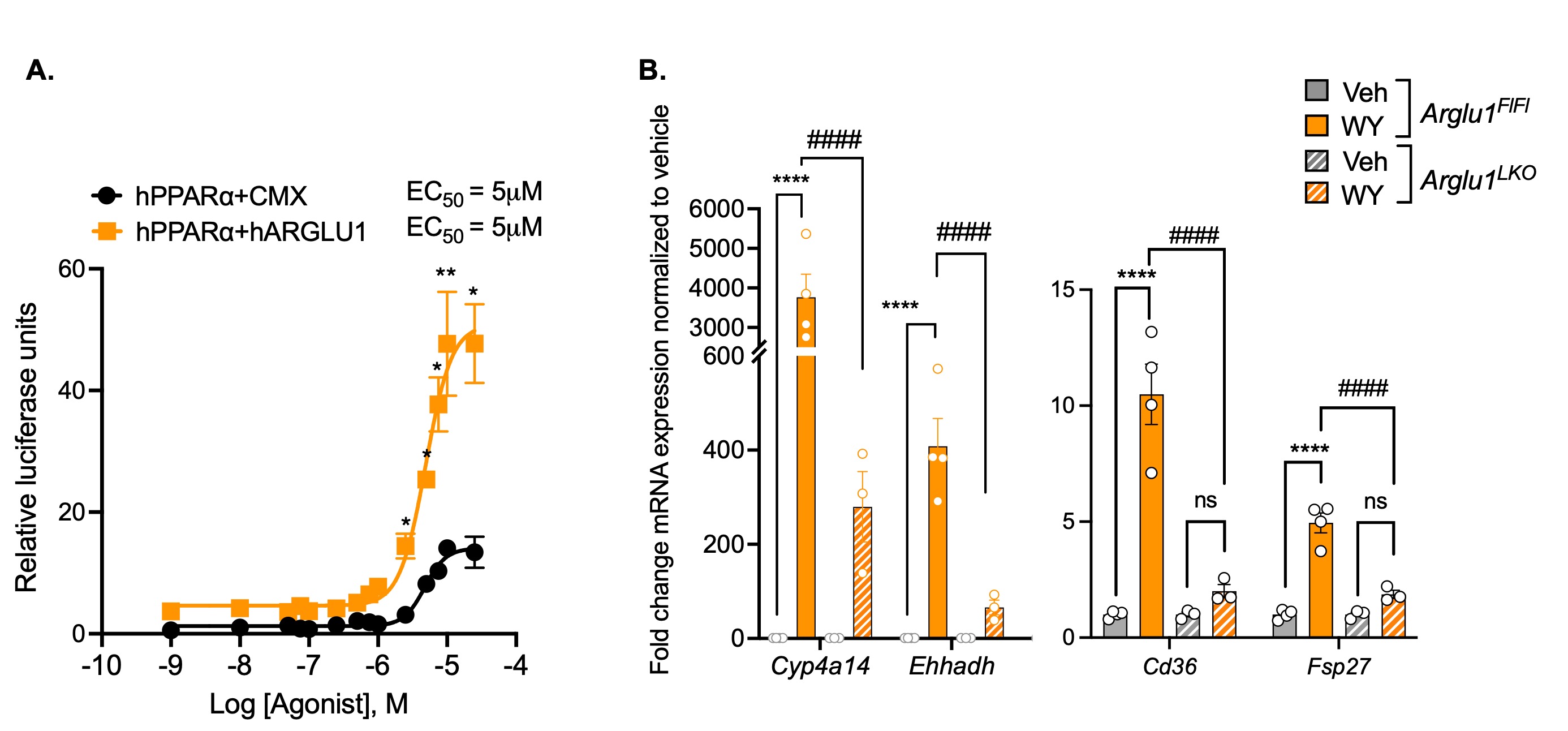
Figure 1: ARGLU1 responsible for maximal PPARα luciferase activity and target gene expression. A) Dose response curves for PPARα activity in HEK293 cells co-transfected with GAL4-hPPARα receptor, UAS-luciferase reporter, and CMX-ARGLU1 or empty-CMX (control); and treated with the PPARα ligand WY14643 at increasing concentrations. Data represent the mean ± SD, N=3. Student’s t-test * P ≤ 0.05, ** P ≤ 0.01, relative to CMX control at the same concentration. B) Ligand-dependent induction of Cyp4a14, Ehhadh, Cd36 and Fsp27 mRNA in Arglu1fl/fl and Arglu1LKO mouse primary hepatocytes treated with 20 μM WY14643 for a 24-hour period. Gene expression measured by qPCR. Data represent the mean ± SEM, N=3-4. Two-way ANOVA, * P ≤ 0.05, ***P ≤ 0.001, **** P ≤ 0.0001 versus respective Veh. # P ≤ 0.05, ## P ≤ 0.01, ### P ≤ 0.001, #### P ≤ 0.0001 versus respective Arglu1LKO group. Data from Cash et al. 2021.
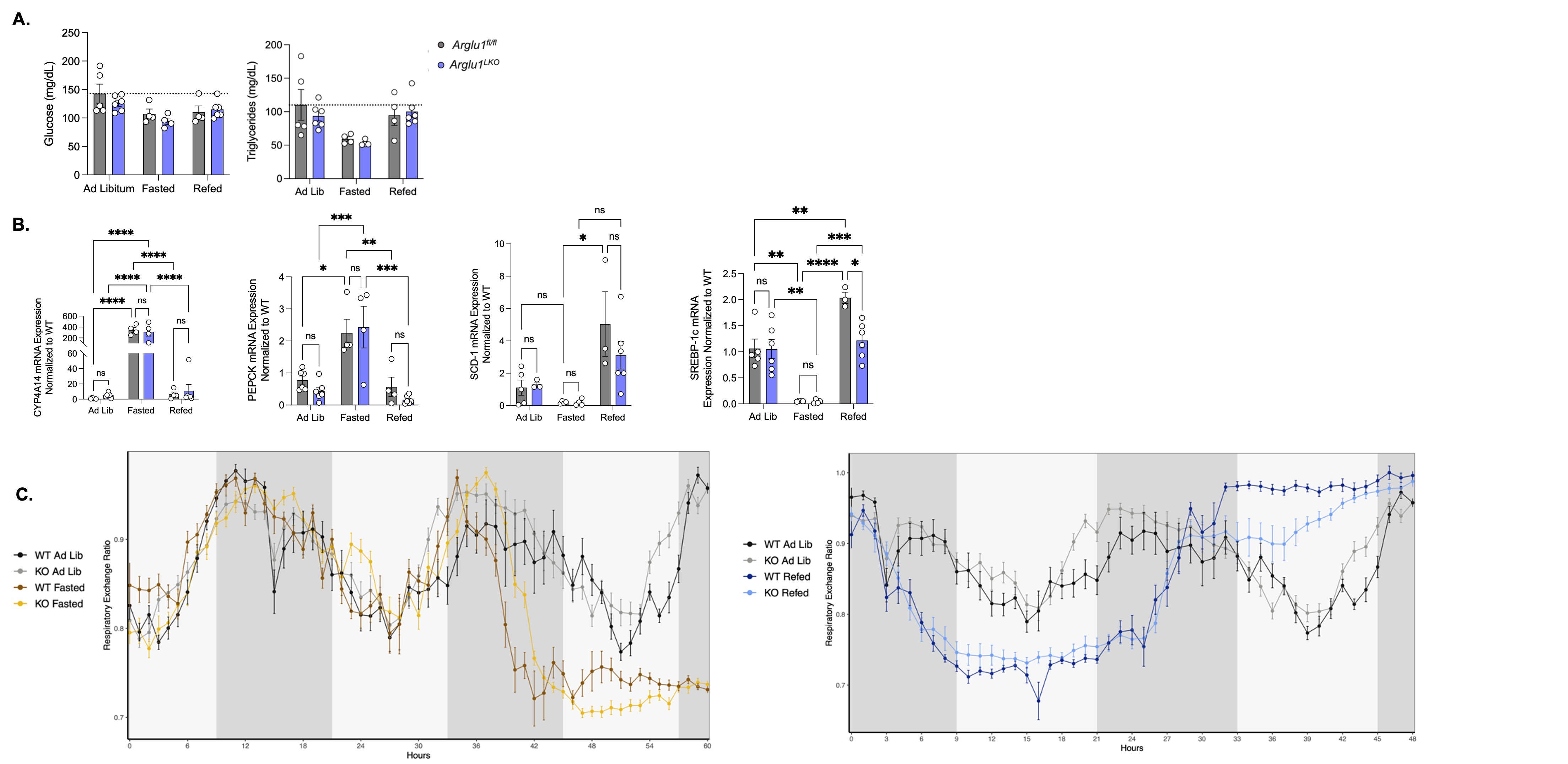
Figure 2. Role of liver Arglu1 in the fasting and refeeding processes. A) Plasma metabolites glucose and triglycerides measured for Arglu1flfl (grey bars) and Arglu1LKO (purple bars) male mice. Two-way ANOVA followed by Newman- Keuls test, no significance found. B) Upon fasting, liver gene expression of Cyp4a14 and Pepck increased, while Scd-1 and Srebp-1c expression decreased, normalized to 36B4 reference gene. Arglu1flfl and Arglu1LKO mice within the refed group exhibited a significant increase in Srebp-1c expression. Data analyzed via two-way ANOVA, *P ≤ 0.05. C) Respiratory exchange ratio for male Arglu1flfl and Arglu1LKO in ad libitum (WT: black, LKO: grey), fasted (WT: brown, LKO: yellow), and refed (WT: dark blue, LKO: light blue) conditions. Raw data was collected for five-minute time intervals, and analyzed using CalR program (CalR Version 1.3, MA). Data represent the mean ± SEM, N=3-4.
.jpg)
Figure 3: Hepatic gene expression from female Arglu1f/lfl and Arglu1LKO mice treated with PPARα agonist. A) Increased hepatic gene expression with PPARα activation from 40 mg/kg WY14643 in Wildtype (WT) Arglu1fl/fl female mice, and dampened hepatic gene expression in Arglu1LKO mice. B) Gene expression that was either unchanged (Ehhadh), or elevated (Fsp27, Cyp4a14) within Arglu1LKO mice compared to Arglu1fl/fl mice upon WY14643 treatment. C) Plasma Glucose (mg/dL) concentrations of Arglu1fl/fl and Arglu1LKO mice following 14-days treatment of either vehicle or WY14643. Data represented as mean ± SEM, N=4-6. Two-way ANOVA followed by Newman-Keuls test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, **** P ≤ 0.0001 versus respective Veh. #P ≤ 0.05, ##P ≤ 0.01, ###P ≤ 0.001, ####P≤ 0.0001 versus respective Arglu1LKO group.
Discovery and Basic Research - Biology
Category: Late Breaking Poster Abstract
(T1230-10-56) Liver-Specific Coactivator ARGLU1 is Required for Selected In Vivo PPARα-Regulated Pathways
Tuesday, October 18, 2022
12:30 PM – 1:30 PM ET
.jpg)
Sarah Cash, BS
PhD Candidate
University of Toronto
Toronto, Ontario, Canada.jpg)
Sarah Cash, BS
PhD Candidate
University of Toronto
Toronto, Ontario, Canada
Presenting Author(s)
Main Author(s)
Purpose: In humans, there are 48 nuclear hormone receptors (NRs) that act as ligand-activated transcription factors to regulate various biological processes, such as metabolism, development, cell proliferation, immune response, central nervous system function, and reproduction. In addition to ligand binding, NR activation is highly controlled through co-regulatory proteins, in which coactivators increase transcription, and corepressors decrease target transcription. The Cummins laboratory discovered a new coactivator protein, arginine and glutamate rich 1 (ARGLU1)1 that significantly increases the activity of the metabolically important peroxisome proliferator-activated receptors (PPARs) (Figure 1)2. PPAR isoforms PPARα, PPARδ and PPARγ, are key modulators of lipid and carbohydrate metabolism and cellular differentiation. Therefore, we will examine the role of ARGLU1 in PPARα-regulated pathways including the fasting/refeeding cycle and under PPARα-specific ligand activation.
Methods: Since mouse knockouts of ARGLU1 were previously shown to be embryonically lethal, our laboratory produced floxed-Arglu1 mice (Arglu1fl/fl) and cross-bred these mice with Albumin-Cre mice to generate liver-specific Arglu1-null mice (Arglu1LKO).1 To determine if ARGLU1 could play a role in PPARα regulated pathways, we performed several in vivo experiments with Arglu1fl/fl and Arglu1LKO mice. When examining fasting and refeeding regulation, mice were housed in Promethion Metabolic Cages (Sable Systems) which measured food and water intake, physical activity, O2 consumption and CO2 production. This allowed the examination on whether changes in fuel utilization and energy expenditure are dependent on the hepatic expression of ARGLU1. The chow feeding conditions were either ad libitum fed, fasted for 24-hours, or fasted for 24 hours and refed for 24-hours. Plasma was collected for metabolite quantification. Hepatic gene expression was also examined for genes important to fasting and refeeding processes. When investigating the importance of liver Arglu1 upon PPARα activation, we treated 14-week-old Arglu1fl/fl and Arglu1LKO mice with 40 mg/kg WY14643 (a PPARα agonist) intraperitoneally for a 14-day period. Quadricep skeletal muscle, liver, intestine and epididymal white adipose tissues were flash frozen at -80°C for gene expression analysis. Liver RNA was isolated and target gene regulation (i.e., Cyp4a14, Ehhadh, Cd36, Fsp27, Fabp2, Pdk4) was examined via RT-qPCR. Plasma metabolites examined include glucose, cholesterol, triglycerides, glycerol, and non-esterified fatty acids.
Results: Results from the fasting and refeeding study demonstrate that ARGLU1 may have an important role in regulating the PPARα response to refeeding. Plasma glucose and triglyceride levels were unchanged between Arglu1fl/fl and Arglu1LKO mice (Figure 2a). Hepatic gene expression demonstrated the expected patterns of gene induction or repression upon fasting; however, there were no changes between Arglu1fl/fl and Arglu1LKO genotypes (Figure 2b). The lack of difference between genotypes in genes responsible for feeding regulation was surprising, as changes in the magnitude of PPARα target gene expression were anticipated based on previous studies performed in primary hepatocytes (Figure 1b). However, there was a significant difference between Arglu1fl/fl and Arglu1LKO refed groups in the gene regulation of Srebp-1c, a lipogenic regulator, which is highly increased upon refeeding, and dampened without ARGLU1 (Figure 2b). The respiratory exchange ratio (RER) followed the expected patterns of 0.9 upon feeding, indicating preferential glucose utilization, and around 0.7 RER with fasting when fat utilization is maximal for Arglu1fl/fl and Arglu1LKO groups (Figure 2c). Liver RNA was isolated from Arglu1fl/fl and Arglu1LKO mice treated with either vehicle or WY14643, and target gene regulation was examined via RT-qPCR. Specific genes involved in fatty acid utilization (Fgf21, Pdk4, Fabp2, Cd36) were dampened in the Arglu1LKO group upon WY14643 treatment (Figure 3a). However, not all genes were decreased, and some important fatty acid metabolic genes (Cyp4a14, Fsp27) were increased in Arglu1LKO mice (Figure 3b). No difference in plasma glucose levels was observed between genotypes (Figure 3c).
Conclusion: We conclude that ARGLU1 impacts select PPAR-regulated lipid metabolic pathways in unanticipated ways. The fasting response was not dramatically impacted; however, mice expressing ARGLU1 showed a tendency towards enhanced carbohydrate utilization on refeeding and warrants further investigation into its role in intermediary metabolism in vivo. Future directions also include examining the role of ARGLU1 in PPAR response to a high-fat diet.
References: 1. Magomedova L, Tiefenbach J, Zilberman E, Le Billan F, Voisin V, Saikali M, Boivin V, Robitaille M, Gueroussov S, Irimia M, Ray D, Patel R, Xu C, Jeyasuria P, Bader GD, Hughes TR, Morris QD, Scott MS, Krause H, Angers S, Blencowe BJ, Cummins CL. ARGLU1 is a transcriptional coactivator and splicing regulator important for stress hormone signaling and development. Nucleic Acids Res. 2019 Apr 8;47(6):2856-2870.
2. Cash SB., Sahin C, & Cummins CL. (2021). The role of liver arginine and glutamate rich 1 in coactivating PPAR nuclear receptor-mediated gene expression. American Association of Pharmaceutical Sciences PharmSci 360. Virtual.
Acknowledgments: We gratefully acknowledge funding for this project from the Canadian Institutes of Health Research.

Figure 1: ARGLU1 responsible for maximal PPARα luciferase activity and target gene expression. A) Dose response curves for PPARα activity in HEK293 cells co-transfected with GAL4-hPPARα receptor, UAS-luciferase reporter, and CMX-ARGLU1 or empty-CMX (control); and treated with the PPARα ligand WY14643 at increasing concentrations. Data represent the mean ± SD, N=3. Student’s t-test * P ≤ 0.05, ** P ≤ 0.01, relative to CMX control at the same concentration. B) Ligand-dependent induction of Cyp4a14, Ehhadh, Cd36 and Fsp27 mRNA in Arglu1fl/fl and Arglu1LKO mouse primary hepatocytes treated with 20 μM WY14643 for a 24-hour period. Gene expression measured by qPCR. Data represent the mean ± SEM, N=3-4. Two-way ANOVA, * P ≤ 0.05, ***P ≤ 0.001, **** P ≤ 0.0001 versus respective Veh. # P ≤ 0.05, ## P ≤ 0.01, ### P ≤ 0.001, #### P ≤ 0.0001 versus respective Arglu1LKO group. Data from Cash et al. 2021.

Figure 2. Role of liver Arglu1 in the fasting and refeeding processes. A) Plasma metabolites glucose and triglycerides measured for Arglu1flfl (grey bars) and Arglu1LKO (purple bars) male mice. Two-way ANOVA followed by Newman- Keuls test, no significance found. B) Upon fasting, liver gene expression of Cyp4a14 and Pepck increased, while Scd-1 and Srebp-1c expression decreased, normalized to 36B4 reference gene. Arglu1flfl and Arglu1LKO mice within the refed group exhibited a significant increase in Srebp-1c expression. Data analyzed via two-way ANOVA, *P ≤ 0.05. C) Respiratory exchange ratio for male Arglu1flfl and Arglu1LKO in ad libitum (WT: black, LKO: grey), fasted (WT: brown, LKO: yellow), and refed (WT: dark blue, LKO: light blue) conditions. Raw data was collected for five-minute time intervals, and analyzed using CalR program (CalR Version 1.3, MA). Data represent the mean ± SEM, N=3-4.
.jpg)
Figure 3: Hepatic gene expression from female Arglu1f/lfl and Arglu1LKO mice treated with PPARα agonist. A) Increased hepatic gene expression with PPARα activation from 40 mg/kg WY14643 in Wildtype (WT) Arglu1fl/fl female mice, and dampened hepatic gene expression in Arglu1LKO mice. B) Gene expression that was either unchanged (Ehhadh), or elevated (Fsp27, Cyp4a14) within Arglu1LKO mice compared to Arglu1fl/fl mice upon WY14643 treatment. C) Plasma Glucose (mg/dL) concentrations of Arglu1fl/fl and Arglu1LKO mice following 14-days treatment of either vehicle or WY14643. Data represented as mean ± SEM, N=4-6. Two-way ANOVA followed by Newman-Keuls test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, **** P ≤ 0.0001 versus respective Veh. #P ≤ 0.05, ##P ≤ 0.01, ###P ≤ 0.001, ####P≤ 0.0001 versus respective Arglu1LKO group.
