Back
Purpose: Broadly neutralizing antibodies (bNAbs) identified from HIV-1 patients are promising as therapeutics or preventions of HIV-1 infection, and they also provide an opportunity to study human humoral immune responses against HIV-1 infection. Previous studies have shown that highly potent bNAbs only develop in a small number of patients over a long course of continuous exposure to the virus infection1,2. Thus, it is important to study how antibody responses change through different times of the infection in these individuals to better understand the shape of cross-reactive antibodies and the maturation process of bNAbs. To profile the dynamic changes of cross-reactivity of human antibody repertoires, here we sequenced the longitudinal antibody repertoires of 2 HIV-1 infected donors with exceptional HIV-1 neutralization breadth, known as CAP292 and CAP312, in a paired heavy:light chain format. We also immortalized these precious human B cell receptor (BCR) libraries in a renewable yeast display format, so that BCRs expressed on the yeast surface as antigen-binding fragments (Fab) would enable precise functional screening of antibody activity against numerous HIV-1 strains and Env antigens.
Methods: Peripheral blood mononuclear cells (PBMCs) from 4 chronologic time points of CAP292 and CAP312 were obtained from our collaborators. CD27+ antigen-experienced memory B cells were isolated through magnetic selection and stimulated in vitro for 5 days before processing. The single B cell immune repertoire sequencing was conducted using an axisymmetric flow-focusing device3. Briefly, a single cell was captured and lysed in a single emulsion droplet and its mRNAs were annealed to the poly(dT) beads. An emulsion-based overlap-extension (OE) RT-PCR reaction was used to join the genes of the heavy chain variable domain (VH) and light chain variable domain (VL) through a linker to form paired VH:VL amplicons. The resulting amplicon libraries were sent for next-generation sequencing (NGS) via Illumina Miseq 2*300. To construct antibody yeast display libraries, VH:VL cDNA libraries were first inserted into the yeast display vector, then digested and ligated with the bidirectional promoter to obtain the final DNA plasmid libraries. Antibody yeast libraries were prepared by transforming the final plasmid libraries into yeast using a high-efficiency yeast electroporation protocol. To induce Fab expression, yeast libraries were cultured in SGDCAA media for 36 hours. On the day of the experiment, yeast cells were stained with anti-FLAG FITC, and Fab-expressing populations (VL+) were sorted through anti-FITC magnetic beads enrichment (MACS). Yeast cells were also stained with R-Phycoerythrin (PE) conjugated BG505 and DU172.17 Env trimers to check for antigen binding.
Results: We successfully amplified paired VH:VL amplicons from the memory B cells of these two donors and submitted the amplicons for NGS sequence analysis. Using established bioinformatics tools, we analyzed the NGS data of these paired heavy and light libraries, and we found that we recovered between 1,000 to 10,000 unique paired VH:VL clusters for each time point sample. We calculated the number of unique VH:VL clusters divided by the number of input cells, which ranged from around 1% to 7.5% (Figure 1). Paired VH:VL amplicon libraries of donor CAP292 were next cloned into the yeast display system for immortalization as natively paired antibody yeast libraries. For each transformation step, we achieved at least a 100-fold higher number of transformants than the number of unique clusters to guarantee full coverage of the paired VH:VL amplicon libraries. Preliminary FACS data showed that CAP292 kappa presort libraries showed around 5% antibody expression, which increased to around 8% after magnetic enrichment of VL+ clones. Three kappa libraries from CAP292 exhibited distinct binding patterns against the HIV-1 Env trimers BG505 SOSIP and DU172.17 (Figure 2).
Conclusion: We successfully captured and sequenced the longitudinal paired VH:VL repertoire from two chronic HIV-1 infected donors, CAP292 and CAP312, with the recovery of around 10,000 and 20,000 unique VH:VL clusters respectively. Paired VH:VL amplicon libraries were further cloned into the yeast display system which allows repetitive functional screening studies to understand the longitudinal evolution of cross-reactive antibodies and bNAbs.
References: 1. Hraber, P. et al. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169 (2014).
2. Liao, H.-X. et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013).
3. McDaniel, J. R., DeKosky, B. J., Tanno, H., Ellington, A. D. & Georgiou, G. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat Protoc 11, 429–442 (2016).
.jpg)
Figure 1. Summary of memory B cell selection, NGS results, and neutralization potency of each time point sample of the two donors. NGS results are shown as the number of combined unique clusters for each isotype of antibody in the generated libraries. A unique VH:VL cluster is a bioinformatic approximation of a distinct antibody lineage; each unique VH:VL cluster can contain multiple distinct antibody sequences or clonal variants.
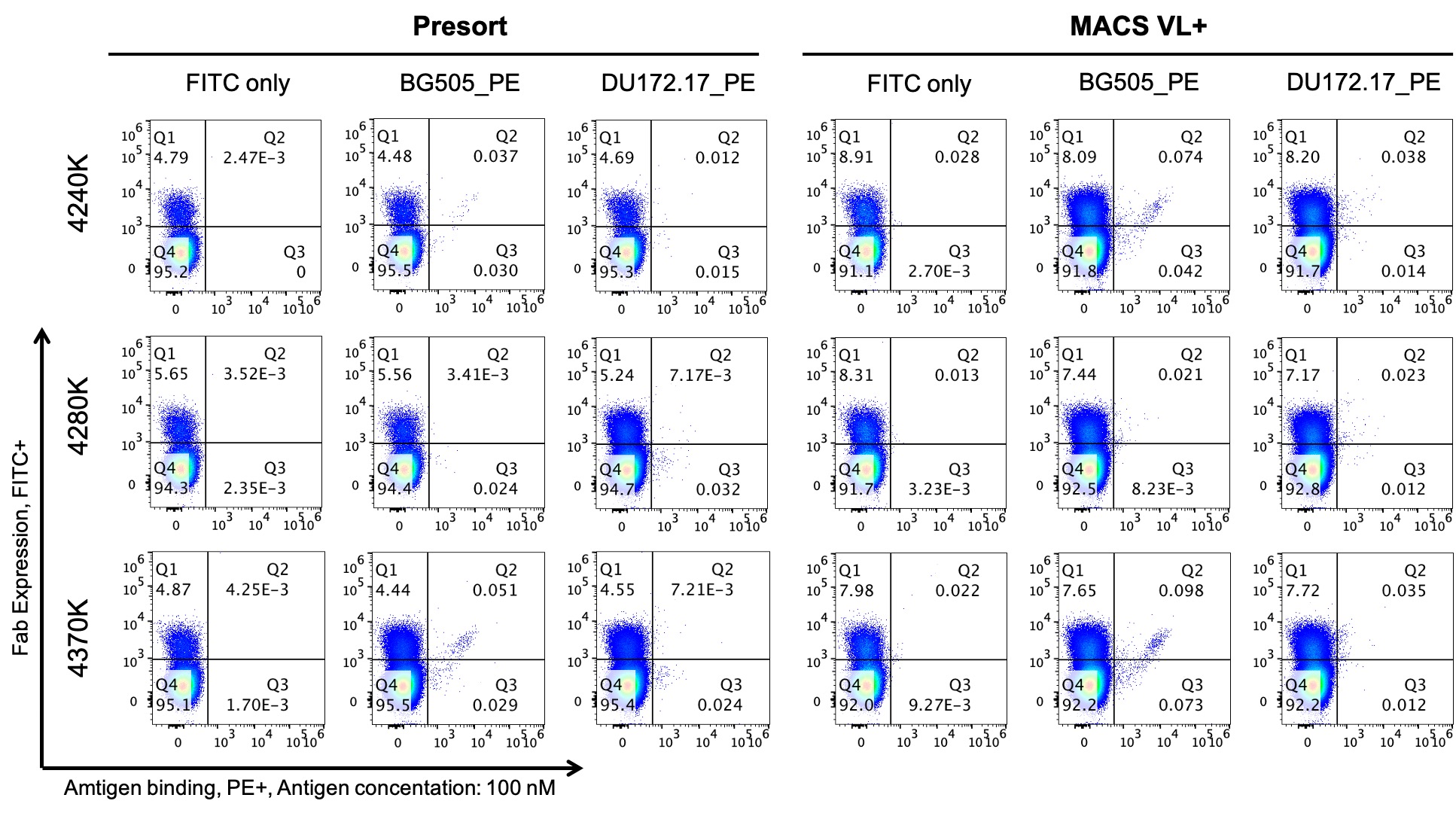
Figure 2. Flow cytometry data showed that the antibody expression level was increased after VL+ enrichment, and different libraries had distinct antigen binding patterns against two HIV-1 Env trimers.
Discovery and Basic Research - Biology
Category: Late Breaking Poster Abstract
(T1030-05-26) Immortalization and Functional Screening of Human Antibody Repertoires from Chronic HIV-1 Infected Patients
Tuesday, October 18, 2022
10:30 AM – 11:30 AM ET
- XP
Xiaoli Pan, MS
University of Kansas
Lawrence, Kansas, United States - XP
Xiaoli Pan, MS
University of Kansas
Lawrence, Kansas, United States
Presenting Author(s)
Main Author(s)
Purpose: Broadly neutralizing antibodies (bNAbs) identified from HIV-1 patients are promising as therapeutics or preventions of HIV-1 infection, and they also provide an opportunity to study human humoral immune responses against HIV-1 infection. Previous studies have shown that highly potent bNAbs only develop in a small number of patients over a long course of continuous exposure to the virus infection1,2. Thus, it is important to study how antibody responses change through different times of the infection in these individuals to better understand the shape of cross-reactive antibodies and the maturation process of bNAbs. To profile the dynamic changes of cross-reactivity of human antibody repertoires, here we sequenced the longitudinal antibody repertoires of 2 HIV-1 infected donors with exceptional HIV-1 neutralization breadth, known as CAP292 and CAP312, in a paired heavy:light chain format. We also immortalized these precious human B cell receptor (BCR) libraries in a renewable yeast display format, so that BCRs expressed on the yeast surface as antigen-binding fragments (Fab) would enable precise functional screening of antibody activity against numerous HIV-1 strains and Env antigens.
Methods: Peripheral blood mononuclear cells (PBMCs) from 4 chronologic time points of CAP292 and CAP312 were obtained from our collaborators. CD27+ antigen-experienced memory B cells were isolated through magnetic selection and stimulated in vitro for 5 days before processing. The single B cell immune repertoire sequencing was conducted using an axisymmetric flow-focusing device3. Briefly, a single cell was captured and lysed in a single emulsion droplet and its mRNAs were annealed to the poly(dT) beads. An emulsion-based overlap-extension (OE) RT-PCR reaction was used to join the genes of the heavy chain variable domain (VH) and light chain variable domain (VL) through a linker to form paired VH:VL amplicons. The resulting amplicon libraries were sent for next-generation sequencing (NGS) via Illumina Miseq 2*300. To construct antibody yeast display libraries, VH:VL cDNA libraries were first inserted into the yeast display vector, then digested and ligated with the bidirectional promoter to obtain the final DNA plasmid libraries. Antibody yeast libraries were prepared by transforming the final plasmid libraries into yeast using a high-efficiency yeast electroporation protocol. To induce Fab expression, yeast libraries were cultured in SGDCAA media for 36 hours. On the day of the experiment, yeast cells were stained with anti-FLAG FITC, and Fab-expressing populations (VL+) were sorted through anti-FITC magnetic beads enrichment (MACS). Yeast cells were also stained with R-Phycoerythrin (PE) conjugated BG505 and DU172.17 Env trimers to check for antigen binding.
Results: We successfully amplified paired VH:VL amplicons from the memory B cells of these two donors and submitted the amplicons for NGS sequence analysis. Using established bioinformatics tools, we analyzed the NGS data of these paired heavy and light libraries, and we found that we recovered between 1,000 to 10,000 unique paired VH:VL clusters for each time point sample. We calculated the number of unique VH:VL clusters divided by the number of input cells, which ranged from around 1% to 7.5% (Figure 1). Paired VH:VL amplicon libraries of donor CAP292 were next cloned into the yeast display system for immortalization as natively paired antibody yeast libraries. For each transformation step, we achieved at least a 100-fold higher number of transformants than the number of unique clusters to guarantee full coverage of the paired VH:VL amplicon libraries. Preliminary FACS data showed that CAP292 kappa presort libraries showed around 5% antibody expression, which increased to around 8% after magnetic enrichment of VL+ clones. Three kappa libraries from CAP292 exhibited distinct binding patterns against the HIV-1 Env trimers BG505 SOSIP and DU172.17 (Figure 2).
Conclusion: We successfully captured and sequenced the longitudinal paired VH:VL repertoire from two chronic HIV-1 infected donors, CAP292 and CAP312, with the recovery of around 10,000 and 20,000 unique VH:VL clusters respectively. Paired VH:VL amplicon libraries were further cloned into the yeast display system which allows repetitive functional screening studies to understand the longitudinal evolution of cross-reactive antibodies and bNAbs.
References: 1. Hraber, P. et al. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169 (2014).
2. Liao, H.-X. et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013).
3. McDaniel, J. R., DeKosky, B. J., Tanno, H., Ellington, A. D. & Georgiou, G. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat Protoc 11, 429–442 (2016).
.jpg)
Figure 1. Summary of memory B cell selection, NGS results, and neutralization potency of each time point sample of the two donors. NGS results are shown as the number of combined unique clusters for each isotype of antibody in the generated libraries. A unique VH:VL cluster is a bioinformatic approximation of a distinct antibody lineage; each unique VH:VL cluster can contain multiple distinct antibody sequences or clonal variants.

Figure 2. Flow cytometry data showed that the antibody expression level was increased after VL+ enrichment, and different libraries had distinct antigen binding patterns against two HIV-1 Env trimers.
