Back
Poster Session C
Genetics, genomics and proteomics
Session: (1118–1149) Genetics, Genomics and Proteomics Poster
1142: The Lupus Risk Allele Associated Xq28 Haplotype Identifies the Molecular Endotype of a Cohort of European SLE Patients with Enhanced IRAK1 Signaling
Sunday, November 13, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- KO
Katherine Owen, PhD
RILITE
Charlottesville, VA, United States
Abstract Poster Presenter(s)
Rheyanna Clemens1, Tyson Dawson1, Katherine Owen2, Amrie Grammer3 and Peter Lipsky1, 1AMPEL BioSolutions, Charlottesville, VA, 2RILITE, Crozet, VA, 3AMPEL LLC, Charlottesville, VA
Background/Purpose: Previous genetic association studies identified a 9 gene region of Xq28 containing IRAK1 as a risk locus for SLE. IRAK1 plays key roles downstream of the endosomal TLRs and IL-1 receptors to activate NF-KB during inflammatory responses. While single nucleotide polymorphisms (SNPs) in IRAK1 have been independently associated with SLE risk, the molecular pathways have not fully delineated the differential impact in lupus patients of different ancestries. Here, we provide a full characterization of the genetic haplotype predictive of a patient endotype characterized by enhanced IRAK1 signaling in European ancestry SLE patients.
Methods: Gene Analysis Tool Kit variant calling was used to identify expression of 150 known Xq28 SNPs in an RNA-seq dataset of SLE patients of European (EA) or Asian (AsA) ancestry. Patient cohorts were determined by SNP expression and visualized via t-SNE plot. Differentially expressed gene (DEG) datasets were created to leverage inter-cohort and intra-ancestral changes in gene expression. DEGs were integrated into protein-protein interaction networks, partitioned with MCODE, and annotated using a variety of bioinformatics tools. Gene set enrichment analysis (GSEA) was utilized to validate overall enrichment of pathways observed with MCODE clustering. For comparison, DEGs derived from IRAK1/2-deficient mice (GSE10765) were clustered and annotated to identify pathways down-regulated in the absence of IRAK1.
Results: Variant calling detected 150 SNPs in the Xq28 region from SLE patients with approximately 33% total coverage. 4 independent cohorts based on SNP expression were grouped by t-SNE. Cohorts 1 and 2 comprised 45% and 33% of subjects, respectively, with cohort 1 being 90% AsA ancestry (AsA-1) and cohort 2 being 90% EA ancestry (EA-2). The AsA-1 haplotype expressed primarily homozygous alternate alleles within AVPR2-TMEM187 and reference alleles elsewhere, whereas the EA-2 haplotype expressed alternative alleles within IRAK1-MECP2 (Figure 1). The smaller cohorts 3 and 4 comprised 21% of all subjects and expressed heterozygous SNPs across the entire region in both ancestries. Pathway analysis of EA-2 DEGs indicated activation of innate and inflammatory immune responses related to TLR signaling that are consistent with IRAK1 up-regulation in this patient cohort. In contrast, AsA-1 had no enrichment of similar pathways. Using the IRAK1/2 KO mouse as reference, we further confirmed that genes down-regulated in the absence of IRAK1 are strong predictors of genes up-regulated in SLE patients of EA ancestry harboring the IRAK1 variant risk allele.
Conclusion: Here, we confirmed expression of previously documented SNPs in Xq28 in SLE patients and discovered 2 distinct haplotypes indicative of patient ancestry. We observed and validated up-regulation of inflammatory pathways downstream of TLR signaling in EA patients expressing alternate alleles in the IRAK1 locus, confirming IRAK1 as a disease-associated gene in these patients. Modulation of IRAK1 signaling could serve as a target for the abrogation of inflammation, predominantly in SLE patients of EA ancestry.
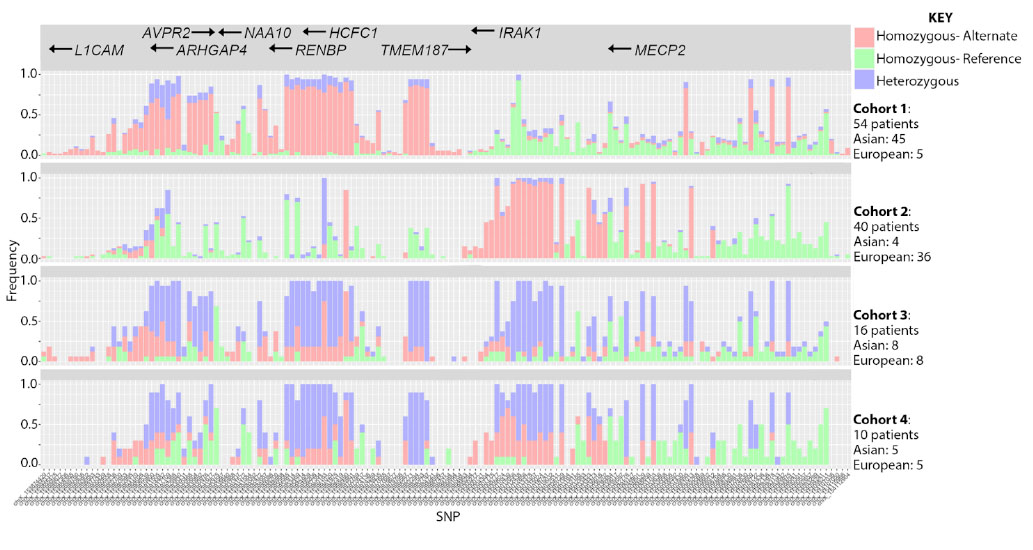 Figure 1: SNP expression, detection and cohort separation annotated with size and ancestral make-up of 4 independent cohorts determined by t-SNE.
Figure 1: SNP expression, detection and cohort separation annotated with size and ancestral make-up of 4 independent cohorts determined by t-SNE.
Disclosures: R. Clemens, None; T. Dawson, None; K. Owen, None; A. Grammer, None; P. Lipsky, None.
Background/Purpose: Previous genetic association studies identified a 9 gene region of Xq28 containing IRAK1 as a risk locus for SLE. IRAK1 plays key roles downstream of the endosomal TLRs and IL-1 receptors to activate NF-KB during inflammatory responses. While single nucleotide polymorphisms (SNPs) in IRAK1 have been independently associated with SLE risk, the molecular pathways have not fully delineated the differential impact in lupus patients of different ancestries. Here, we provide a full characterization of the genetic haplotype predictive of a patient endotype characterized by enhanced IRAK1 signaling in European ancestry SLE patients.
Methods: Gene Analysis Tool Kit variant calling was used to identify expression of 150 known Xq28 SNPs in an RNA-seq dataset of SLE patients of European (EA) or Asian (AsA) ancestry. Patient cohorts were determined by SNP expression and visualized via t-SNE plot. Differentially expressed gene (DEG) datasets were created to leverage inter-cohort and intra-ancestral changes in gene expression. DEGs were integrated into protein-protein interaction networks, partitioned with MCODE, and annotated using a variety of bioinformatics tools. Gene set enrichment analysis (GSEA) was utilized to validate overall enrichment of pathways observed with MCODE clustering. For comparison, DEGs derived from IRAK1/2-deficient mice (GSE10765) were clustered and annotated to identify pathways down-regulated in the absence of IRAK1.
Results: Variant calling detected 150 SNPs in the Xq28 region from SLE patients with approximately 33% total coverage. 4 independent cohorts based on SNP expression were grouped by t-SNE. Cohorts 1 and 2 comprised 45% and 33% of subjects, respectively, with cohort 1 being 90% AsA ancestry (AsA-1) and cohort 2 being 90% EA ancestry (EA-2). The AsA-1 haplotype expressed primarily homozygous alternate alleles within AVPR2-TMEM187 and reference alleles elsewhere, whereas the EA-2 haplotype expressed alternative alleles within IRAK1-MECP2 (Figure 1). The smaller cohorts 3 and 4 comprised 21% of all subjects and expressed heterozygous SNPs across the entire region in both ancestries. Pathway analysis of EA-2 DEGs indicated activation of innate and inflammatory immune responses related to TLR signaling that are consistent with IRAK1 up-regulation in this patient cohort. In contrast, AsA-1 had no enrichment of similar pathways. Using the IRAK1/2 KO mouse as reference, we further confirmed that genes down-regulated in the absence of IRAK1 are strong predictors of genes up-regulated in SLE patients of EA ancestry harboring the IRAK1 variant risk allele.
Conclusion: Here, we confirmed expression of previously documented SNPs in Xq28 in SLE patients and discovered 2 distinct haplotypes indicative of patient ancestry. We observed and validated up-regulation of inflammatory pathways downstream of TLR signaling in EA patients expressing alternate alleles in the IRAK1 locus, confirming IRAK1 as a disease-associated gene in these patients. Modulation of IRAK1 signaling could serve as a target for the abrogation of inflammation, predominantly in SLE patients of EA ancestry.
 Figure 1: SNP expression, detection and cohort separation annotated with size and ancestral make-up of 4 independent cohorts determined by t-SNE.
Figure 1: SNP expression, detection and cohort separation annotated with size and ancestral make-up of 4 independent cohorts determined by t-SNE.Disclosures: R. Clemens, None; T. Dawson, None; K. Owen, None; A. Grammer, None; P. Lipsky, None.

