Back
Ignite Talk
Session: Ignite Session 8A
1139: Molecular Pathways Identified from Risk Alleles Identify Mechanistic Differences in Systemic Lupus Erythematosus Patients of East Asian and European Ancestry
Monday, November 14, 2022
2:05 PM – 2:10 PM Eastern Time
Location: Northern Liberties Stage
- KO
Katherine Owen, PhD
RILITE
Charlottesville, VA, United StatesDisclosure: Disclosure information not submitted.
Ignite Speaker(s)
Katherine Owen1, Kristy Bell2, Andrew Price3, Prathyusha Bachali4, Hannah Ainsworth5, Miranda Marion6, Timothy Howard5, Carl Langefeld7, Nan Shen8, Jinoos Yazdany9, Maria Dall'Era10, Amrie Grammer11 and Peter Lipsky3, 1RILITE, Crozet, VA, 2AMPEL BioSolutions LLC, Charlottesville, VA, 3AMPEL BioSolutions, Charlottesville, VA, 4AMPEL BioSolutions, Redmond, WA, 5Wake Forest School of Medicine, Winston-Salem, NC, 6Wake Forest University, Winston-Salem, NC, 7Wake Forest School of Medicine, Winston Salem, NC, 8Shanghai Jiang Tong University School of Medicine, Shanghai, China, 9UCSF, San Francisco, CA, 10University of California, Division of Rheumatology, San Francisco, CA, 11AMPEL LLC, Charlottesville, VA
Background/Purpose: SLE is a multi-organ autoimmune disorder with a prominent genetic component. Individuals of Asian-Ancestry (AsA) disproportionately experience more severe SLE compared to individuals of European-Ancestry (EA), including increased kidney involvement and tissue damage. Whereas some of this variation may be accounted for by environmental and/or socioeconomic factors, the possibility that risk alleles differentially expressed within patients of AsA ancestry influence the molecular mechanisms involved in SLE pathogenesis and contribute to enhanced clinical severity has not been explored.
Methods: SLE Immunochip-based association studies identified single nucleotide polymorphisms (SNPs) significantly associated with SLE in EA (6748 cases; 11516 controls) and AsA (2485 cases and 3947 controls from Koreans (KR), Han Chinese (HC) and Malaysian Chinese (MC)). To map causal SNPs to genes, we leveraged the relationship between SNPs and genes in coding regions (C-SNP), eQTL status (E-SNP), transcription factors (T-SNP), and proximity (P-SNP). Protein-protein interaction networks were generated by STRING, visualized in Cytoscape, partitioned with MCODE and annotated using Biologically Informed Gene Clustering (BIG-C), Immune (I)-Scope, Ingenuity Pathway Analysis (IPA) and EnrichR. Gene set variation analysis (GSVA) was used to estimate the variation of pre-defined informative gene sets in patient and control samples of gene expression datasets.
Results: We identified 3,066 ancestry-specific and 384 trans-ancestry SNP-linked genes associated with SLE. Genetic associations were examined using connectivity mapping, and gene signatures based on predicted biological pathways were used to interrogate gene expression datasets. SLE-associated pathways in AsA patients included elevated oxidative stress, altered metabolism and mitochondrial dysfunction, whereas SLE-associated pathways in EA patients included a robust interferon response (type I and II) related to enhanced cytosolic nucleic acid sensing and signaling (Figure 1). An independent dataset derived from summary genome-wide association data in an AsA cohort identified similar molecular pathways. Gene expression data from AsA SLE patients corroborated the molecular pathways and dominant cell types predicted by SNP associations.
Conclusion: The SNP-associated predicted genes and gene expression profiles outlined here implicate similarities and also fundamental differences in lupus molecular pathways operative in EA and AsA ancestral populations. Our findings indicate that certain pathways may be enriched in one ancestral population over another, but those pathways may not be active within every patient of a given ancestry and may be active in patients from other ancestries. Systems bioinformatics and assessment using gene signature enrichment analyses revealed alterations in cellular metabolism and cell stress signatures that may be more prevalent in patients of Asian ancestry. Identifying molecular pathways predicted by ancestry specific genetic SLE risk analyses may help to disentangle the population differences in clinical severity that impact AsA and EA individuals with SLE and guide targeted therapies (Figure 1).
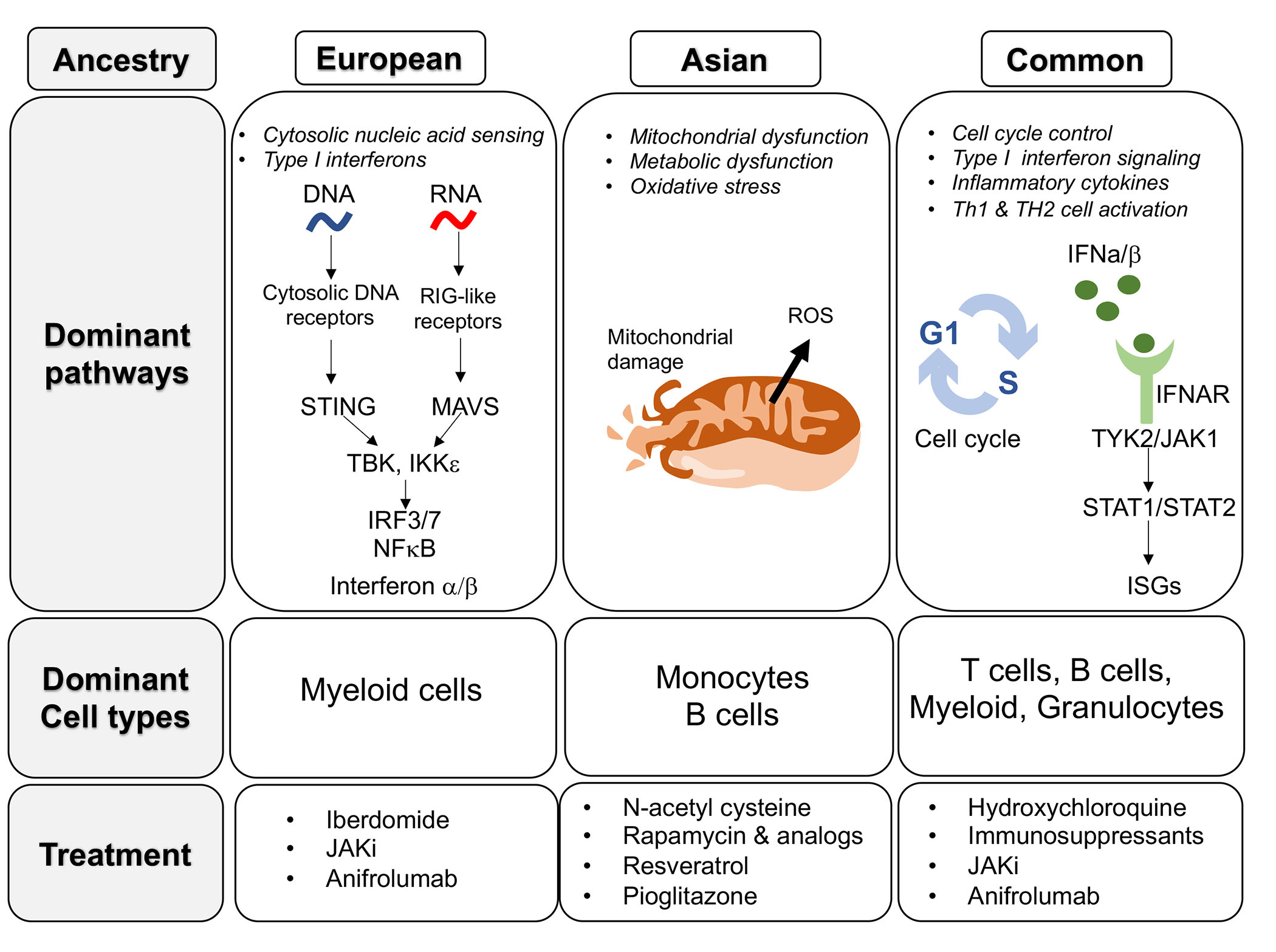 Figure 1. Summary of lupus molecular pathways, dominant cell types and potential treatment options in EA and AsA ancestral populations.
Figure 1. Summary of lupus molecular pathways, dominant cell types and potential treatment options in EA and AsA ancestral populations.
Disclosures: K. Owen, None; K. Bell, None; A. Price, None; P. Bachali, None; H. Ainsworth, None; M. Marion, None; T. Howard, None; C. Langefeld, None; N. Shen, None; J. Yazdany, AstraZeneca, Gilead, Bristol-Myers Squibb(BMS), Aurinia, Astra Zeneca, Pfizer; M. Dall'Era, GlaxoSmithKline (GSK), AstraZeneca, Aurinia, BMS, Amgen; A. Grammer, None; P. Lipsky, None.
Background/Purpose: SLE is a multi-organ autoimmune disorder with a prominent genetic component. Individuals of Asian-Ancestry (AsA) disproportionately experience more severe SLE compared to individuals of European-Ancestry (EA), including increased kidney involvement and tissue damage. Whereas some of this variation may be accounted for by environmental and/or socioeconomic factors, the possibility that risk alleles differentially expressed within patients of AsA ancestry influence the molecular mechanisms involved in SLE pathogenesis and contribute to enhanced clinical severity has not been explored.
Methods: SLE Immunochip-based association studies identified single nucleotide polymorphisms (SNPs) significantly associated with SLE in EA (6748 cases; 11516 controls) and AsA (2485 cases and 3947 controls from Koreans (KR), Han Chinese (HC) and Malaysian Chinese (MC)). To map causal SNPs to genes, we leveraged the relationship between SNPs and genes in coding regions (C-SNP), eQTL status (E-SNP), transcription factors (T-SNP), and proximity (P-SNP). Protein-protein interaction networks were generated by STRING, visualized in Cytoscape, partitioned with MCODE and annotated using Biologically Informed Gene Clustering (BIG-C), Immune (I)-Scope, Ingenuity Pathway Analysis (IPA) and EnrichR. Gene set variation analysis (GSVA) was used to estimate the variation of pre-defined informative gene sets in patient and control samples of gene expression datasets.
Results: We identified 3,066 ancestry-specific and 384 trans-ancestry SNP-linked genes associated with SLE. Genetic associations were examined using connectivity mapping, and gene signatures based on predicted biological pathways were used to interrogate gene expression datasets. SLE-associated pathways in AsA patients included elevated oxidative stress, altered metabolism and mitochondrial dysfunction, whereas SLE-associated pathways in EA patients included a robust interferon response (type I and II) related to enhanced cytosolic nucleic acid sensing and signaling (Figure 1). An independent dataset derived from summary genome-wide association data in an AsA cohort identified similar molecular pathways. Gene expression data from AsA SLE patients corroborated the molecular pathways and dominant cell types predicted by SNP associations.
Conclusion: The SNP-associated predicted genes and gene expression profiles outlined here implicate similarities and also fundamental differences in lupus molecular pathways operative in EA and AsA ancestral populations. Our findings indicate that certain pathways may be enriched in one ancestral population over another, but those pathways may not be active within every patient of a given ancestry and may be active in patients from other ancestries. Systems bioinformatics and assessment using gene signature enrichment analyses revealed alterations in cellular metabolism and cell stress signatures that may be more prevalent in patients of Asian ancestry. Identifying molecular pathways predicted by ancestry specific genetic SLE risk analyses may help to disentangle the population differences in clinical severity that impact AsA and EA individuals with SLE and guide targeted therapies (Figure 1).
 Figure 1. Summary of lupus molecular pathways, dominant cell types and potential treatment options in EA and AsA ancestral populations.
Figure 1. Summary of lupus molecular pathways, dominant cell types and potential treatment options in EA and AsA ancestral populations.Disclosures: K. Owen, None; K. Bell, None; A. Price, None; P. Bachali, None; H. Ainsworth, None; M. Marion, None; T. Howard, None; C. Langefeld, None; N. Shen, None; J. Yazdany, AstraZeneca, Gilead, Bristol-Myers Squibb(BMS), Aurinia, Astra Zeneca, Pfizer; M. Dall'Era, GlaxoSmithKline (GSK), AstraZeneca, Aurinia, BMS, Amgen; A. Grammer, None; P. Lipsky, None.

