Back
Introduction: To develop an automated machine learning model to characterize Prostate Imaging Reporting and Data System (PIRADS) lesions using biparametric (bp) magnetic resonance imaging (MRI).
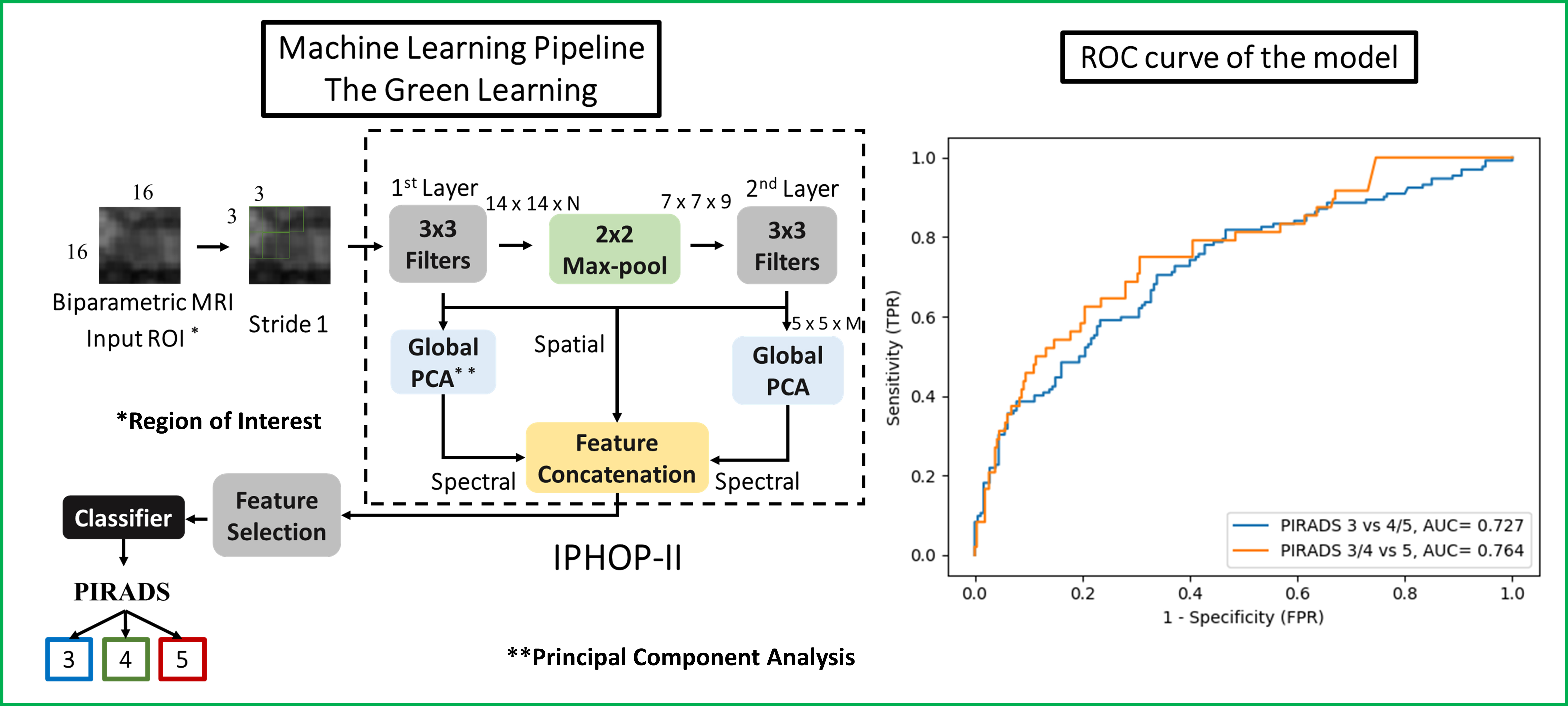
Methods: Consecutive men who underwent 3T prostate MRI (T2-weighted [T2WI], diffusion-weighted images [DWI], and apparent diffusion coefficient [ADC]) followed by prostate biopsy (PBx) were identified from a PBx database (IRB# HS-13-00663). MRI were acquired and interpreted according to PIRADS v2 or v2.1 by experienced radiologists. The PIRADS 3-5 lesions were manually segmented on T2WI, high-b-value (b=1000) DWI, and ADC. Men with single lesion (PIRADS=3) were included. A novel Green Learning framework with a lightweight model and explainable feature extraction process was used (Figure). A data-driven automatic and unsupervised process, namely IP-HOPII, by cascading layers of local-to-global Principal Component Analysis on the input voxels was applied. A spatial-spectral representation was derived of the region of interest. Then a feature selection module was employed to filter out the noisy spectral dimensions. Finally, the best features were fed to the Extreme Gradient Boosting classifier. The performances of classifying PIRADS were assessed by concordance rate for three-class classification and receiver operating characteristic (ROC) analysis for binary classification. The sensitivity and specificity were determined at the Youden index.
Results: Overall, 259 patients (205 for training and 54 for validation) with single lesion were included: PIRADS 3, 4, and 5, were 152 (59%), 70 (27%), and 37 (14%), respectively. The concordance rate between PIRADS and the model was 60% for PZ and 67% for TZ lesions. For the PZ, the area under the ROC curve (AUC) to characterize PIRADS were 0.73 for 3 vs 4-5, and 0.76 for 3-4 vs 5, respectively. The sensitivity and specificity were 75% and 69% to classify PIRADS =4 lesions, and 78% and 71% for PIRADS 5 lesions. For the TZ, the AUC for the model was 0.69 for PIRADS 3 vs 4-5, and 0.64 for PIRADS 3-4 vs 5, respectively. The sensitivity and specificity were 62% and 73% to classify PIRADS =4 lesions, and 60% and 59% for PIRADS 5 lesions.
Conclusions: Machine learning model following the Green Learning paradigm can accurately characterize lesions on prostate bpMRI. SOURCE OF
Funding: None.
Moderated Poster Session
Session: MP55: Prostate Cancer: Detection & Screening III
MP55-18: A Novel Machine Learning Framework to Automated Characterize Prostate Imaging Reporting and Data System (PIRADS) on MRI.
Sunday, April 30, 2023
9:30 AM – 11:30 AM CST
Location: S403
- GC
Poster Presenter(s)
Introduction: To develop an automated machine learning model to characterize Prostate Imaging Reporting and Data System (PIRADS) lesions using biparametric (bp) magnetic resonance imaging (MRI).
Methods: Consecutive men who underwent 3T prostate MRI (T2-weighted [T2WI], diffusion-weighted images [DWI], and apparent diffusion coefficient [ADC]) followed by prostate biopsy (PBx) were identified from a PBx database (IRB# HS-13-00663). MRI were acquired and interpreted according to PIRADS v2 or v2.1 by experienced radiologists. The PIRADS 3-5 lesions were manually segmented on T2WI, high-b-value (b=1000) DWI, and ADC. Men with single lesion (PIRADS=3) were included. A novel Green Learning framework with a lightweight model and explainable feature extraction process was used (Figure). A data-driven automatic and unsupervised process, namely IP-HOPII, by cascading layers of local-to-global Principal Component Analysis on the input voxels was applied. A spatial-spectral representation was derived of the region of interest. Then a feature selection module was employed to filter out the noisy spectral dimensions. Finally, the best features were fed to the Extreme Gradient Boosting classifier. The performances of classifying PIRADS were assessed by concordance rate for three-class classification and receiver operating characteristic (ROC) analysis for binary classification. The sensitivity and specificity were determined at the Youden index.
Results: Overall, 259 patients (205 for training and 54 for validation) with single lesion were included: PIRADS 3, 4, and 5, were 152 (59%), 70 (27%), and 37 (14%), respectively. The concordance rate between PIRADS and the model was 60% for PZ and 67% for TZ lesions. For the PZ, the area under the ROC curve (AUC) to characterize PIRADS were 0.73 for 3 vs 4-5, and 0.76 for 3-4 vs 5, respectively. The sensitivity and specificity were 75% and 69% to classify PIRADS =4 lesions, and 78% and 71% for PIRADS 5 lesions. For the TZ, the AUC for the model was 0.69 for PIRADS 3 vs 4-5, and 0.64 for PIRADS 3-4 vs 5, respectively. The sensitivity and specificity were 62% and 73% to classify PIRADS =4 lesions, and 60% and 59% for PIRADS 5 lesions.
Conclusions: Machine learning model following the Green Learning paradigm can accurately characterize lesions on prostate bpMRI. SOURCE OF
Funding: None.

