Back
Introduction: To develop a novel machine learning framework for automated detection of clinically significant prostate cancer (CSPCa) on Prostate Imaging Reporting & Data System (PIRADS) lesions confirmed by magnetic resonance imaging (MRI) informed target biopsy (TB).
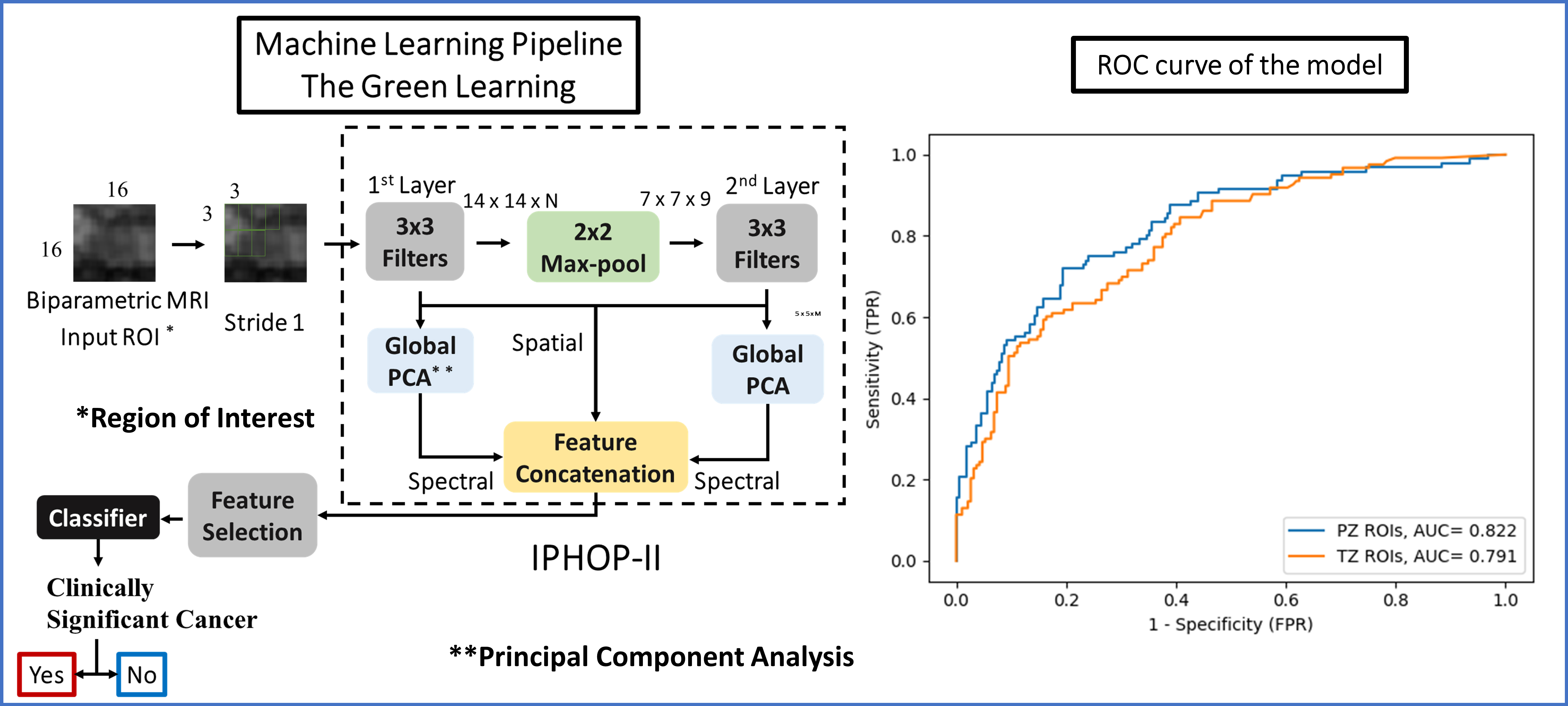
Methods: From our prostate biopsy (PBx) database (IRB# HS-13-00663), consecutive men who underwent 3T multiparametric MRI (T2-weighted [T2W], diffusion-weighted images [DWI], apparent diffusion coefficient [ADC] and Dynamic Contrast Enhanced) followed by MRI-informed TB were identified. MRI was acquired and interpreted according to PIRADS v2 or v2.1 by experienced radiologists. The PIRADS 3-5 lesions were manually segmented on T2W, high-b-value (b=1000) DWI, and ADC. Men with single lesion on MRI (PIRADS=3) were included in the study. CSPCa was defined as Grade Group = 2 on TB. Independent modules were used for peripheral zones (PZ) and transition zones (TZ) regions of interest (ROI). For feature extraction and ROI classification, we adopt the Green Learning (GL) paradigm, and specifically the IPHOP-II feature extraction process (Figure). This offers advantages of lightweight model size and an explainable and transparent feature extraction. The performances of the GL model were assessed by the receiver operating characteristic (ROC) analysis.
Results: A total of 205 MRIs were used for training the GL model and it was validated on 54 MRIs. The PIRADS 3, 4 and 5 distribution was 152, 70 and 37 for PZ, and 84, 24, 15 for TZ, respectively. The CSPCa detection for PIRADS 3, 4 and 5 was 16%, 46% and 68% for PZ, and 13%, 29%, and 53%, for TZ, respectively. The area under the ROC curve (AUC) for the PZ on validation set was: 0.82 for overall; 0.71 for PIRADS 3; 0.80 for PIRADS 4; and 0.87 for PIRADS 5, respectively. The error rate was 27% for PIRADS 3; 18% for PIRADS 4; and 12% for PIRADS 5. The AUC for the TZ on validating set were: 0.79 for overall; 0.69 for PIRADS 3; 0.81 for PIRADS 4; and 0.86 for PIRADS 5, respectively. The error rate was 29% for PIRADS 3; 19% for PIRADS 4; and 13% for PIRADS 5.
Conclusions: We developed a novel machine learning framework, the Green Learning, to detect CSPCa according to PIRADS stratification. This new tool assists with the proper prostatic lesions classification with improved accuracy to the current radiology performance. SOURCE OF
Funding: None.
Moderated Poster Session
Session: MP55: Prostate Cancer: Detection & Screening III
MP55-20: A Novel Machine Learning Framework for Automated Detection of Prostate Cancer Lesions Confirmed on MRI-Informed Target Biopsy.
Sunday, April 30, 2023
9:30 AM – 11:30 AM CST
Location: S403

Masatomo Kaneko, MD, PhD (he/him/his)
University of Southern California
Poster Presenter(s)
Introduction: To develop a novel machine learning framework for automated detection of clinically significant prostate cancer (CSPCa) on Prostate Imaging Reporting & Data System (PIRADS) lesions confirmed by magnetic resonance imaging (MRI) informed target biopsy (TB).
Methods: From our prostate biopsy (PBx) database (IRB# HS-13-00663), consecutive men who underwent 3T multiparametric MRI (T2-weighted [T2W], diffusion-weighted images [DWI], apparent diffusion coefficient [ADC] and Dynamic Contrast Enhanced) followed by MRI-informed TB were identified. MRI was acquired and interpreted according to PIRADS v2 or v2.1 by experienced radiologists. The PIRADS 3-5 lesions were manually segmented on T2W, high-b-value (b=1000) DWI, and ADC. Men with single lesion on MRI (PIRADS=3) were included in the study. CSPCa was defined as Grade Group = 2 on TB. Independent modules were used for peripheral zones (PZ) and transition zones (TZ) regions of interest (ROI). For feature extraction and ROI classification, we adopt the Green Learning (GL) paradigm, and specifically the IPHOP-II feature extraction process (Figure). This offers advantages of lightweight model size and an explainable and transparent feature extraction. The performances of the GL model were assessed by the receiver operating characteristic (ROC) analysis.
Results: A total of 205 MRIs were used for training the GL model and it was validated on 54 MRIs. The PIRADS 3, 4 and 5 distribution was 152, 70 and 37 for PZ, and 84, 24, 15 for TZ, respectively. The CSPCa detection for PIRADS 3, 4 and 5 was 16%, 46% and 68% for PZ, and 13%, 29%, and 53%, for TZ, respectively. The area under the ROC curve (AUC) for the PZ on validation set was: 0.82 for overall; 0.71 for PIRADS 3; 0.80 for PIRADS 4; and 0.87 for PIRADS 5, respectively. The error rate was 27% for PIRADS 3; 18% for PIRADS 4; and 12% for PIRADS 5. The AUC for the TZ on validating set were: 0.79 for overall; 0.69 for PIRADS 3; 0.81 for PIRADS 4; and 0.86 for PIRADS 5, respectively. The error rate was 29% for PIRADS 3; 19% for PIRADS 4; and 13% for PIRADS 5.
Conclusions: We developed a novel machine learning framework, the Green Learning, to detect CSPCa according to PIRADS stratification. This new tool assists with the proper prostatic lesions classification with improved accuracy to the current radiology performance. SOURCE OF
Funding: None.

