Back
Introduction: Radiation therapy (RT), including radioligand therapy, is widely used for the treatment of prostate cancer (PCa). Although the majority of PCa can be effectively treated with RT, approximately 50% of men with high risk PCa will recur. A number of mechanisms, including DNA repair, cell cycle and apoptotic pathways can influence cellular sensitivity to radiation and chemotherapy. Despite recent progress in genomic profiling of PCa, RT-related biomarkers for improving outcome and reducing toxicity remain unknown. Here, we used genome-wide CRISPR screens to systematically map radiosensitivity and gene interactions and to identify novel therapeutic targets in PCa.
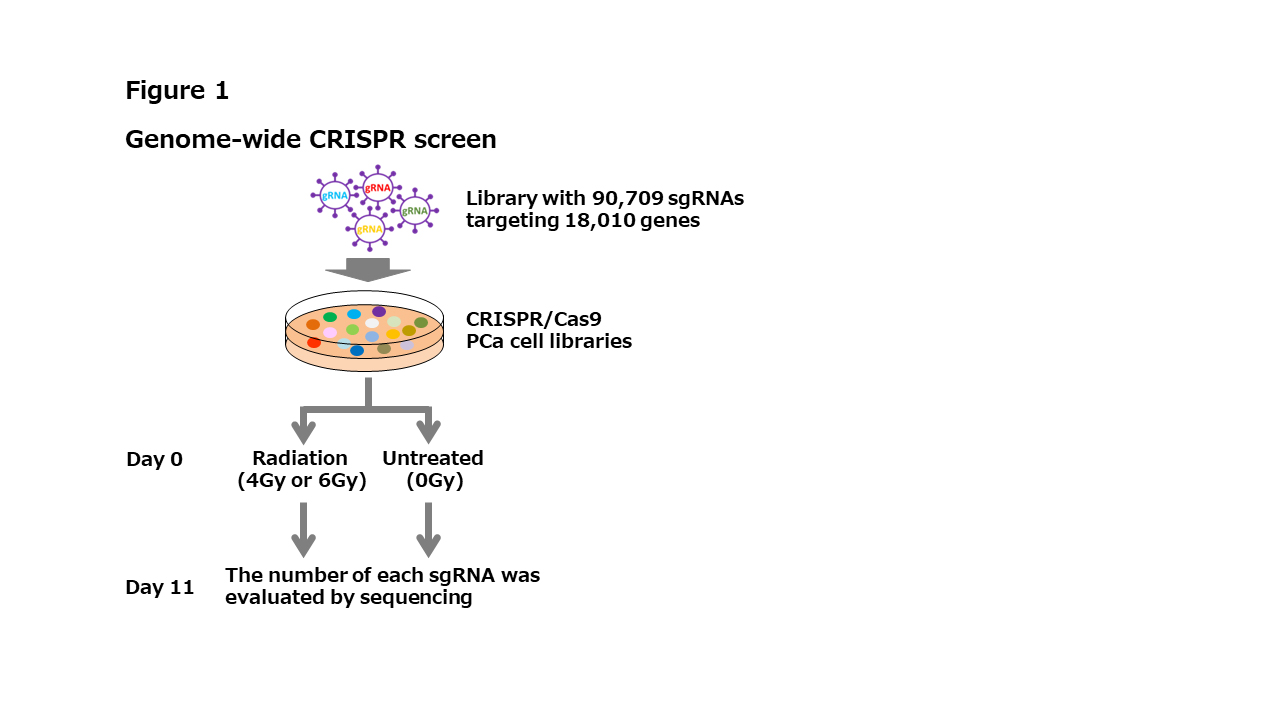
Methods: PCa cells lines, DU145, 22Rv1, and LNCaP, were used to establish CRISPR/Cas9 cell libraries. CRISPR library contained 90,709 sgRNAs targeting 18,010 genes. CRISPR/Cas9 cell libraries were irradiated (4 or 6 Gy) or untreated (control), and cultured for 11 days. The number of each sgRNA was evaluated by sequencing and compared to controls (Figure 1). sgRNA dropoff indicates radiosensitizing genes and sgRNA enrich indicates radioprotective genes. Robust Rank Aggregation (RRA) score was utilized as an estimation for robust identification of CRISPR-screen hits. Top ranked radiosensitizing genes were selected by RRA score in CRISPR screen by three PCa cell lines.
Results: CRISPR screens showed that radiosensitizing genes abundantly contain DNA repair pathway genes, while radioprotective genes contain cell cycle and apoptosis-related genes. Among PCa-related genes, ATM, NBN and PTEN were identified as radiosensitizing genes, while CHEK2 and TP53 were identified as radioprotective genes. The top ranked radiosensitizing genes were identified by CRISPR screens of three PCa cell lines, and validated by each gene knockout using multiple cell lines. The interaction of androgen receptor signaling and radiosensitizing genes was demonstrated using androgen suppression and testosterone addition in LNCaP cells.
Conclusions: Functional CRISPR screens revealed genome-wide RT responses in PCa, providing new information for the development of personalized RT. SOURCE OF
Funding: This research was supported by the Japan Society for the Promotion of Science KAKENHI, grant number 20K18090.
Moderated Poster Session
Session: MP20: Prostate Cancer: Basic Research & Pathophysiology I
MP20-09: Genome-wide CRISPR screens identify novel target genes which alter prostate cancer sensitivity to radiation therapy
Friday, April 28, 2023
3:30 PM – 5:30 PM CST
Location: S403

Koji Hatano (he/him/his)
Osaka University
Poster Presenter(s)
Introduction: Radiation therapy (RT), including radioligand therapy, is widely used for the treatment of prostate cancer (PCa). Although the majority of PCa can be effectively treated with RT, approximately 50% of men with high risk PCa will recur. A number of mechanisms, including DNA repair, cell cycle and apoptotic pathways can influence cellular sensitivity to radiation and chemotherapy. Despite recent progress in genomic profiling of PCa, RT-related biomarkers for improving outcome and reducing toxicity remain unknown. Here, we used genome-wide CRISPR screens to systematically map radiosensitivity and gene interactions and to identify novel therapeutic targets in PCa.
Methods: PCa cells lines, DU145, 22Rv1, and LNCaP, were used to establish CRISPR/Cas9 cell libraries. CRISPR library contained 90,709 sgRNAs targeting 18,010 genes. CRISPR/Cas9 cell libraries were irradiated (4 or 6 Gy) or untreated (control), and cultured for 11 days. The number of each sgRNA was evaluated by sequencing and compared to controls (Figure 1). sgRNA dropoff indicates radiosensitizing genes and sgRNA enrich indicates radioprotective genes. Robust Rank Aggregation (RRA) score was utilized as an estimation for robust identification of CRISPR-screen hits. Top ranked radiosensitizing genes were selected by RRA score in CRISPR screen by three PCa cell lines.
Results: CRISPR screens showed that radiosensitizing genes abundantly contain DNA repair pathway genes, while radioprotective genes contain cell cycle and apoptosis-related genes. Among PCa-related genes, ATM, NBN and PTEN were identified as radiosensitizing genes, while CHEK2 and TP53 were identified as radioprotective genes. The top ranked radiosensitizing genes were identified by CRISPR screens of three PCa cell lines, and validated by each gene knockout using multiple cell lines. The interaction of androgen receptor signaling and radiosensitizing genes was demonstrated using androgen suppression and testosterone addition in LNCaP cells.
Conclusions: Functional CRISPR screens revealed genome-wide RT responses in PCa, providing new information for the development of personalized RT. SOURCE OF
Funding: This research was supported by the Japan Society for the Promotion of Science KAKENHI, grant number 20K18090.

