Back
Introduction: To develop an automated machine learning (ML) model to segment the prostate gland, the peripheral zone (PZ), and the transition zone (TZ) using magnetic resonance image (MRI) and to create a web-based software interface for annotation.
Methods: Consecutive men who underwent prostate MRI followed by prostate biopsy (PBx) were identified from our PBx database (IRB# HS-13-00663). The 3T MRI was performed according to Prostate Imaging-Reporting and Data System (PIRADS) v2 or v2.1. The T2-weighted (T2W) images were manually segmented into the whole prostate, PZ, and TZ by experienced radiologist and urologist. A novel two-stage automatic Green Learning (GL) based machine learning model was designed, which is a novel non-deep learning method. The first stage segments the prostate gland and the second stage zooms into the prostate area to delineate TZ and PZ. Both stages share a lightweight feed-forward encoder-decoder GL system. Included accessions were split for 5-fold cross-validation. The volumes were calculated according to the number of pixels/voxels. The model performance for automated prostate segmentation was evaluated by Dice scores and Pearson correlation coefficients. The web-based software interface was designed and implemented for users to interact with the AI annotation model and make necessary adjustments.
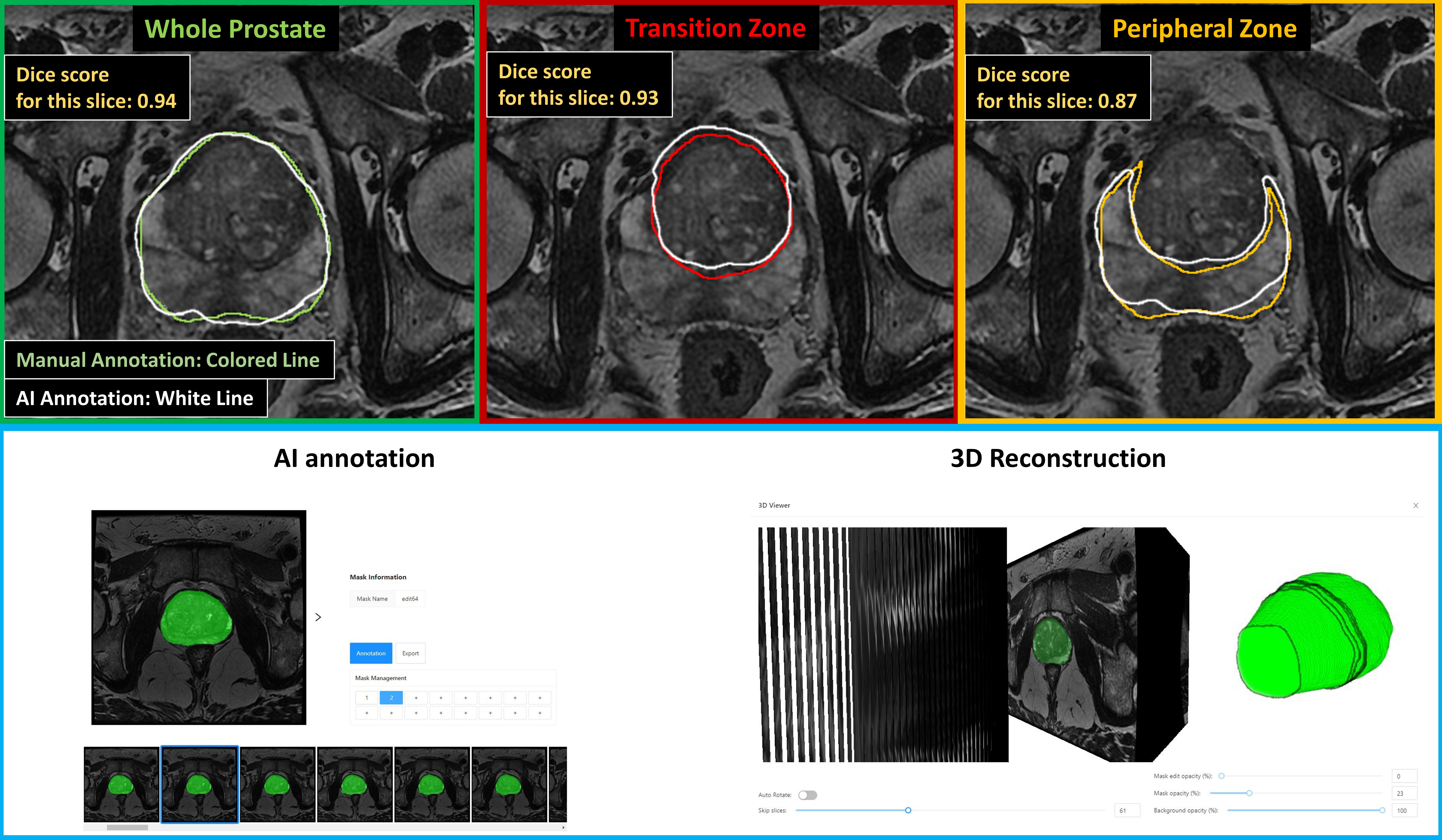
Results: A total of 119 patients (19992 T2W images) met the inclusion criteria (Figure). Using the training dataset of 95 MRIs, a ML model for whole prostate, PZ, and TZ segmentation was constructed. The mean Dice scores for whole prostate, PZ, and TZ were 0.85, 0.62 and 0.81, respectively. The Pearson correlation coefficient for volumes of whole prostate, PZ, and TZ segmentation were 0.92 (p < 0.01), 0.62 (p < 0.01), and 0.93 (p < 0.01), respectively. The web-based software interface takes a mean of 90sec for prostate segmentation with 168 slices. The platform supports DICOM series upload, image preview, image modification, 3-dimensional preview, and annotation mask export, from any device without migrating data.
Conclusions: A lightweight feed-forward encoder-decoder model based on Green Learning can precisely segment the whole prostate, PZ and TZ. This is available on a user-friendly software interface. SOURCE OF
Funding: None.
Moderated Poster Session
Session: MP09: Imaging/Radiology: Uroradiology I
MP09-05: Automated Prostate Gland and Prostate Zones Segmentation Using a Novel MRI-Based Machine Learning Framework and Creation of Software Interface for Users Annotation
Friday, April 28, 2023
9:30 AM – 11:30 AM CST
Location: S404C

Masatomo Kaneko, MD, PhD (he/him/his)
University of Southern California
Poster Presenter(s)
Introduction: To develop an automated machine learning (ML) model to segment the prostate gland, the peripheral zone (PZ), and the transition zone (TZ) using magnetic resonance image (MRI) and to create a web-based software interface for annotation.
Methods: Consecutive men who underwent prostate MRI followed by prostate biopsy (PBx) were identified from our PBx database (IRB# HS-13-00663). The 3T MRI was performed according to Prostate Imaging-Reporting and Data System (PIRADS) v2 or v2.1. The T2-weighted (T2W) images were manually segmented into the whole prostate, PZ, and TZ by experienced radiologist and urologist. A novel two-stage automatic Green Learning (GL) based machine learning model was designed, which is a novel non-deep learning method. The first stage segments the prostate gland and the second stage zooms into the prostate area to delineate TZ and PZ. Both stages share a lightweight feed-forward encoder-decoder GL system. Included accessions were split for 5-fold cross-validation. The volumes were calculated according to the number of pixels/voxels. The model performance for automated prostate segmentation was evaluated by Dice scores and Pearson correlation coefficients. The web-based software interface was designed and implemented for users to interact with the AI annotation model and make necessary adjustments.
Results: A total of 119 patients (19992 T2W images) met the inclusion criteria (Figure). Using the training dataset of 95 MRIs, a ML model for whole prostate, PZ, and TZ segmentation was constructed. The mean Dice scores for whole prostate, PZ, and TZ were 0.85, 0.62 and 0.81, respectively. The Pearson correlation coefficient for volumes of whole prostate, PZ, and TZ segmentation were 0.92 (p < 0.01), 0.62 (p < 0.01), and 0.93 (p < 0.01), respectively. The web-based software interface takes a mean of 90sec for prostate segmentation with 168 slices. The platform supports DICOM series upload, image preview, image modification, 3-dimensional preview, and annotation mask export, from any device without migrating data.
Conclusions: A lightweight feed-forward encoder-decoder model based on Green Learning can precisely segment the whole prostate, PZ and TZ. This is available on a user-friendly software interface. SOURCE OF
Funding: None.

