Back
Introduction: Germline mutations in DNA-damage repair (DDR) genes such as BRCA2 have been associated with prostate cancer (PC) risk but has not been thoroughly evaluated for metastatic prostate cancer (mPC) in Asian men. This study attempts to evaluate frequency of DDR mutations in the largest cohort of Koreans.
Methods: We recruited 340 patients with mPC unselected for family history of cancer and compared to 495 controls. Whole genome sequencing was applied to assess germline pathogenic/likely pathogenic variants (PVs/LPVs) in 26 DDR genes and HOXB13, including 7 genes (ATM, BRCA1/2, CHEK2, BRIP1, PALB2, and NBN) associated with hereditary PC. Comparisons to published Caucasian and Japanese cohorts were performed.
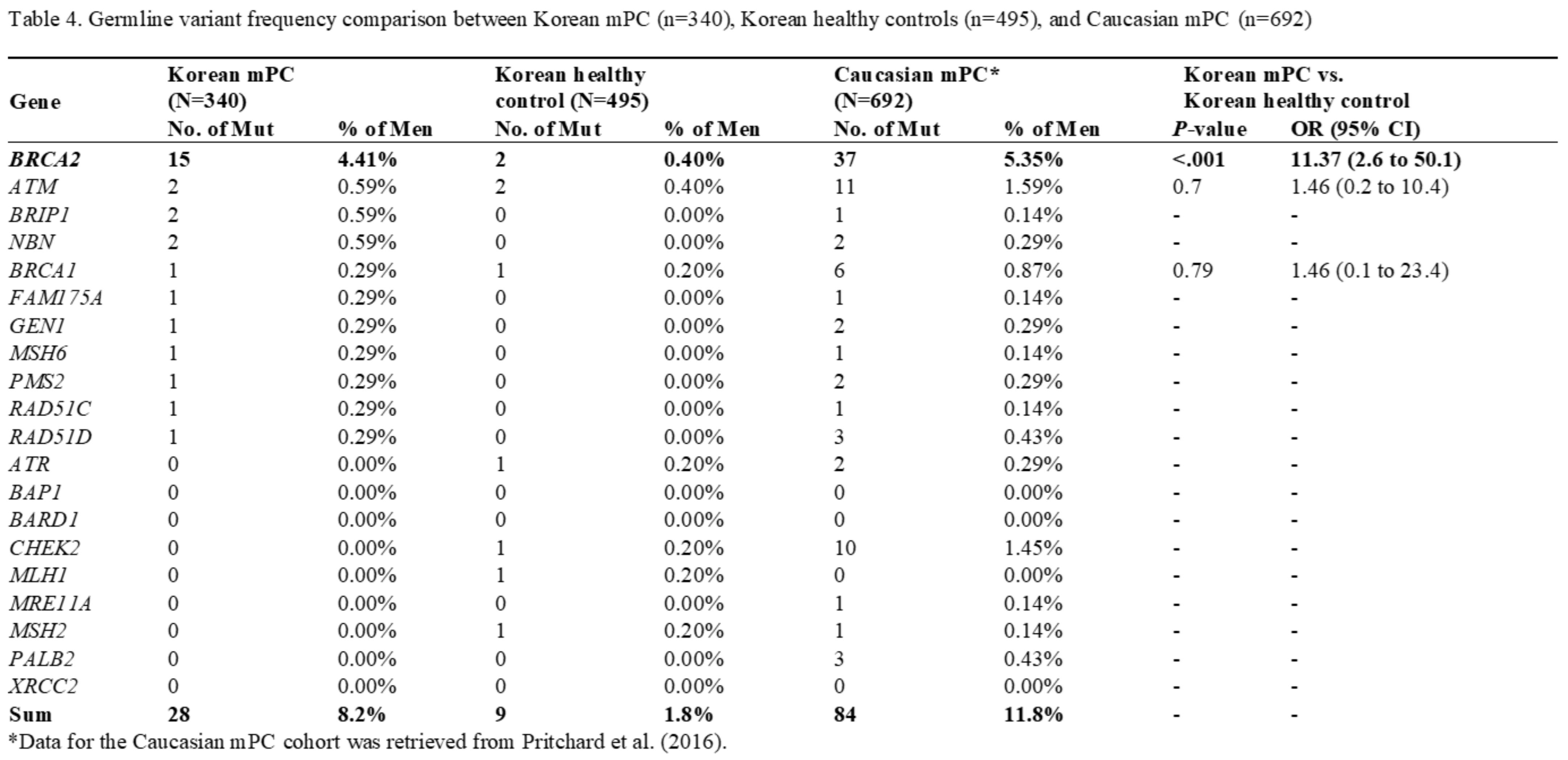
Results: Total of 28 PVs/LPVs were identified in 30 (8.5%) patients; mutations were found in 13 genes, including BRCA2 (15 men [4.4%]), ATM (2 men [0.6%]), NBN (2 men [0.6%], and BRIP1 (2 men [0.6%]). Only one patient had HOXB13 mutation (0.3%). Compared with the germline variant frequency of previously reported mPC studies (8.5% and 11.8%), a slightly lower or similar rate was found in Korean mPC, but notably differed from Caucasian and geographically similar Japanese cohorts. PVs/LPVs in DDR genes tended to increase gradually with higher Gleason Scores (GS 7, 7.1%; GS 8, 7.5%; GS 9-10, 9.9%).
Conclusions: We successfully identified 26 DDR and HOXB13-related deleterious variants in 30 Korean men with aggressive mPC. Germline PV/LPV profiles show comparable frequency of overall carriers (8.5%) but distinctly different distribution when contrasted to previous studies in European and even geographically nearby Japanese cohorts, with BRCA2 playing a dominant role. These results illustrate further evidence for population-based, ethnic-specific analyses for genetic testing as well as highlighting the potential differences that exist even in common East Asian ancestry. These findings may further emphasize the importance of genomic background in Korean PC. Future combinatory efforts must be made in larger, multiethnic trials to identify PVs of variable importance depending on ancestry. SOURCE OF
Funding: This study was supported by the SNUBH Research foundation and KUOS.
Moderated Poster Session
Session: MP17: Prostate Cancer: Markers
MP17-17: Germline mutational carrier frequency analysis in Korean metastatic prostate cancer : a large cohort study
Friday, April 28, 2023
1:00 PM – 3:00 PM CST
Location: S504
- JN
Poster Presenter(s)
Introduction: Germline mutations in DNA-damage repair (DDR) genes such as BRCA2 have been associated with prostate cancer (PC) risk but has not been thoroughly evaluated for metastatic prostate cancer (mPC) in Asian men. This study attempts to evaluate frequency of DDR mutations in the largest cohort of Koreans.
Methods: We recruited 340 patients with mPC unselected for family history of cancer and compared to 495 controls. Whole genome sequencing was applied to assess germline pathogenic/likely pathogenic variants (PVs/LPVs) in 26 DDR genes and HOXB13, including 7 genes (ATM, BRCA1/2, CHEK2, BRIP1, PALB2, and NBN) associated with hereditary PC. Comparisons to published Caucasian and Japanese cohorts were performed.
Results: Total of 28 PVs/LPVs were identified in 30 (8.5%) patients; mutations were found in 13 genes, including BRCA2 (15 men [4.4%]), ATM (2 men [0.6%]), NBN (2 men [0.6%], and BRIP1 (2 men [0.6%]). Only one patient had HOXB13 mutation (0.3%). Compared with the germline variant frequency of previously reported mPC studies (8.5% and 11.8%), a slightly lower or similar rate was found in Korean mPC, but notably differed from Caucasian and geographically similar Japanese cohorts. PVs/LPVs in DDR genes tended to increase gradually with higher Gleason Scores (GS 7, 7.1%; GS 8, 7.5%; GS 9-10, 9.9%).
Conclusions: We successfully identified 26 DDR and HOXB13-related deleterious variants in 30 Korean men with aggressive mPC. Germline PV/LPV profiles show comparable frequency of overall carriers (8.5%) but distinctly different distribution when contrasted to previous studies in European and even geographically nearby Japanese cohorts, with BRCA2 playing a dominant role. These results illustrate further evidence for population-based, ethnic-specific analyses for genetic testing as well as highlighting the potential differences that exist even in common East Asian ancestry. These findings may further emphasize the importance of genomic background in Korean PC. Future combinatory efforts must be made in larger, multiethnic trials to identify PVs of variable importance depending on ancestry. SOURCE OF
Funding: This study was supported by the SNUBH Research foundation and KUOS.

