Back
Introduction: A urine test capable of detecting upper tract urothelial carcinoma (UTUC) that provides diagnostic and prognostic information might aid diagnosis and improve risk stratification, which are major clinical challenges in UTUC. Urinary comprehensive genomic profiling (uCGP) via next-generation sequencing with the CLIA-validated UroAmp assay (Convergent Genomics) was developed to identify mutations, diagnose disease, assess molecular grade and stage, and predict recurrence risk. Here we verify that a urine-based uCGP approach can identify lesions in UTUC patients and we compare these genomic profiles to subjects with bladder cancer.
Methods: uCGP was performed on 69 specimens from individuals with de novo pathology-confirmed UC of the bladder (UCB) and 12 urine specimens from individuals with de novo pathology-confirmed UTUC. Urine DNA was sequenced, comprehensively profiled across 60 genes, and disease classification was predicted by a machine learned algorithm. Differential mutational signals of UTUC and UCB were compared through odds ratio (OR) analysis with statistical significance ascribed by Fisher’s exact test.
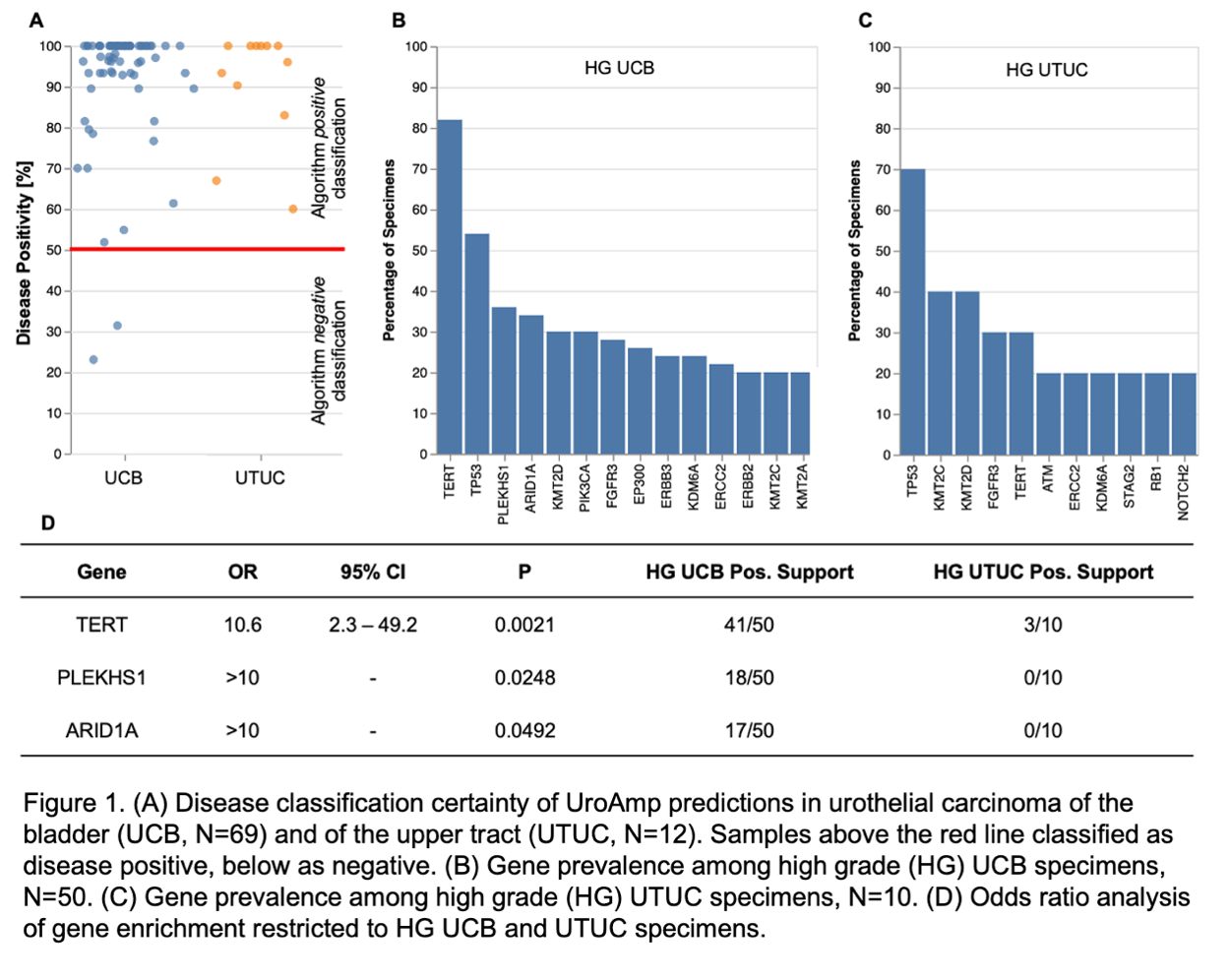
Results: Among UTUC patients, grade distribution was 83% HG, 17% LG; stage distribution was 25% Ta, 25% T1, 8% T2, 8% T3, 8% T4, 25% unknown; among UCB patients grade distribution was 70% HG, 30% LG; stage distribution was 45% Ta, 22% T1, 20% T2, 7% T4, 4% Tis, 2% unknown. uCGP correctly classified 100% of UTUC subjects and 97% (67/69) of UCB subjects as disease positive. UTUC lesions had a high prevalence of TP53 mutations (70%) and frequently demonstrated genomic instability with large scale aneuploidy (40%), loss of heterozygosity (30%), ERCC2 amplification (30%), and CDKN2A loss (10%). Additional recurrent mutations included KMT2C (40%), KMT2D (40%), FGFR3 (30%), and TERT (30%). OR analysis revealed significant enrichment of promoter mutations in TERT (OR=10.6) among high grade (HG) UCB lesions compared to HG UTUC. Further, mutations in ARID1A, PLEKHS1, were solely observed in HG UCB lesions.
Conclusions: uCGP is a quantitative diagnostic with the ability to profile genomic lesions in UTUC and UCB. While further study is needed, distinct mutational patterns in UTUC could potentially serve to identify urologic origins of urine-based disease signals. SOURCE OF
Funding: Research support by Convergent Genomics
Podium Session
Session: PD46: Bladder Cancer: Upper Tract Transitional Cell Carcinoma II
PD46-07: Urinary comprehensive genomic profiling aids in detection and risk prognosis of upper tract urothelial carcinoma: a case-controlled cohort study
Monday, May 1, 2023
2:00 PM – 2:10 PM CST
Location: S404A
- PY
Podium Presenter(s)
Introduction: A urine test capable of detecting upper tract urothelial carcinoma (UTUC) that provides diagnostic and prognostic information might aid diagnosis and improve risk stratification, which are major clinical challenges in UTUC. Urinary comprehensive genomic profiling (uCGP) via next-generation sequencing with the CLIA-validated UroAmp assay (Convergent Genomics) was developed to identify mutations, diagnose disease, assess molecular grade and stage, and predict recurrence risk. Here we verify that a urine-based uCGP approach can identify lesions in UTUC patients and we compare these genomic profiles to subjects with bladder cancer.
Methods: uCGP was performed on 69 specimens from individuals with de novo pathology-confirmed UC of the bladder (UCB) and 12 urine specimens from individuals with de novo pathology-confirmed UTUC. Urine DNA was sequenced, comprehensively profiled across 60 genes, and disease classification was predicted by a machine learned algorithm. Differential mutational signals of UTUC and UCB were compared through odds ratio (OR) analysis with statistical significance ascribed by Fisher’s exact test.
Results: Among UTUC patients, grade distribution was 83% HG, 17% LG; stage distribution was 25% Ta, 25% T1, 8% T2, 8% T3, 8% T4, 25% unknown; among UCB patients grade distribution was 70% HG, 30% LG; stage distribution was 45% Ta, 22% T1, 20% T2, 7% T4, 4% Tis, 2% unknown. uCGP correctly classified 100% of UTUC subjects and 97% (67/69) of UCB subjects as disease positive. UTUC lesions had a high prevalence of TP53 mutations (70%) and frequently demonstrated genomic instability with large scale aneuploidy (40%), loss of heterozygosity (30%), ERCC2 amplification (30%), and CDKN2A loss (10%). Additional recurrent mutations included KMT2C (40%), KMT2D (40%), FGFR3 (30%), and TERT (30%). OR analysis revealed significant enrichment of promoter mutations in TERT (OR=10.6) among high grade (HG) UCB lesions compared to HG UTUC. Further, mutations in ARID1A, PLEKHS1, were solely observed in HG UCB lesions.
Conclusions: uCGP is a quantitative diagnostic with the ability to profile genomic lesions in UTUC and UCB. While further study is needed, distinct mutational patterns in UTUC could potentially serve to identify urologic origins of urine-based disease signals. SOURCE OF
Funding: Research support by Convergent Genomics

