Back
Introduction: Systemic neoadjuvant chemotherapy (NAC) may apply selective pressure on tumors and shape cancer evolution. In bladder cancer, intratumor genetic heterogeneity can help predict response to NAC, with RB1, ERCC2, ATM, and FANCC mutations significantly associated with a positive response to NAC. Molecular studies have shown upper tract urothelial carcinoma (UTUC) to be genetically distinct from bladder cancer with a significantly worse response to NAC, yet still harboring significant intratumor heterogeneity. This study aims to assess the effect of NAC in selecting chemotherapy-resistant mutations among the intratumor heterogeneity in UTUC.
Methods: We identified patients with UTUC who underwent platinum-based NAC followed by nephroureterectomy treated at Memorial Sloan Kettering Cancer Center. Patients with complete response to NAC were excluded. Pre-NAC biopsy and post-NAC nephroureterectomy samples underwent next generation targeted gene sequencing through MSK-IMPACT. Mutational frequency among the 505 tested genes was compared in the pre-and post- treatment cohorts to assess for significant changes (p < 0.05).
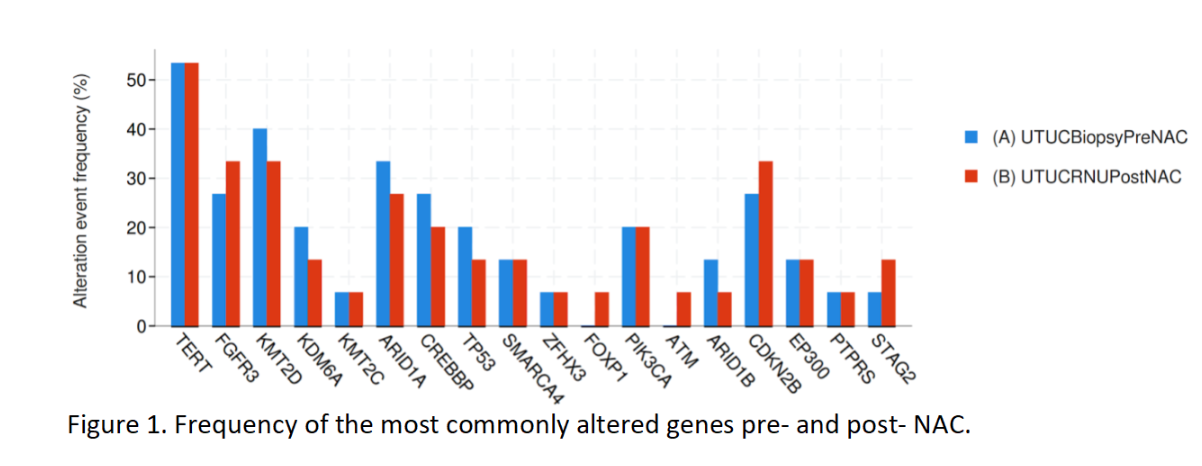
Results: We identified 15 patients with UTUC that had an incomplete response/progression of disease after NAC with gemcitabine/cisplatin as determined by final pathology at radical nephroureterectomy. The median age at the time of NAC was 60 (IQR 55-70). The most common mutations are seen in Figure 1. The most common mutated genes in pre-NAC UTUC biopsy were TERT (53.3%), KMT2D (40.0%), and ARID1A (33.3%). Post-chemotherapy, the most common mutations were TERT (56.3%), FGFR3 (31.3%), and KMT2D (31.3%). There were no significant differences between the frequency of mutations pre- and post-NAC. There was no decrease in the frequency of RB1, ERCC2, ATM, and FANCC post-NAC.
Conclusions: Administration of NAC did not select for specific resistant mutations in the global tumor landscape. It is possible that changes to the tumor microenvironment, sub-clonal expansion, or differences in expression may be identified through RNA based studies. SOURCE OF
Funding: N/A
Podium Session
Session: PD46: Bladder Cancer: Upper Tract Transitional Cell Carcinoma II
PD46-11: Assessing the Role of Neoadjuvant Chemotherapy in Upper Tract Urothelial Carcinoma Evolution
Monday, May 1, 2023
2:40 PM – 2:50 PM CST
Location: S404A
- ND
Podium Presenter(s)
Introduction: Systemic neoadjuvant chemotherapy (NAC) may apply selective pressure on tumors and shape cancer evolution. In bladder cancer, intratumor genetic heterogeneity can help predict response to NAC, with RB1, ERCC2, ATM, and FANCC mutations significantly associated with a positive response to NAC. Molecular studies have shown upper tract urothelial carcinoma (UTUC) to be genetically distinct from bladder cancer with a significantly worse response to NAC, yet still harboring significant intratumor heterogeneity. This study aims to assess the effect of NAC in selecting chemotherapy-resistant mutations among the intratumor heterogeneity in UTUC.
Methods: We identified patients with UTUC who underwent platinum-based NAC followed by nephroureterectomy treated at Memorial Sloan Kettering Cancer Center. Patients with complete response to NAC were excluded. Pre-NAC biopsy and post-NAC nephroureterectomy samples underwent next generation targeted gene sequencing through MSK-IMPACT. Mutational frequency among the 505 tested genes was compared in the pre-and post- treatment cohorts to assess for significant changes (p < 0.05).
Results: We identified 15 patients with UTUC that had an incomplete response/progression of disease after NAC with gemcitabine/cisplatin as determined by final pathology at radical nephroureterectomy. The median age at the time of NAC was 60 (IQR 55-70). The most common mutations are seen in Figure 1. The most common mutated genes in pre-NAC UTUC biopsy were TERT (53.3%), KMT2D (40.0%), and ARID1A (33.3%). Post-chemotherapy, the most common mutations were TERT (56.3%), FGFR3 (31.3%), and KMT2D (31.3%). There were no significant differences between the frequency of mutations pre- and post-NAC. There was no decrease in the frequency of RB1, ERCC2, ATM, and FANCC post-NAC.
Conclusions: Administration of NAC did not select for specific resistant mutations in the global tumor landscape. It is possible that changes to the tumor microenvironment, sub-clonal expansion, or differences in expression may be identified through RNA based studies. SOURCE OF
Funding: N/A

