Back
Introduction: Pathogenic microbes can induce cellular dysfunction and alters immune response to therapy. Taking advantage of single-cell RNA-sequencing (scRNA-seq), we aim to assess the transcriptomic features of the single cell level to identify intracellular pathogens in patients with high grade bladder cancer.
Methods: CellRanger and Alevin were utilized to extract cell barcodes and unique molecular identifiers. Low complexity reads <20 was considered negative, and PCR duplicates were removed. Seuret Findmarkers, scIntegrate, and scDensity Plot were utilized to normalize the data, cluster the cells, and calculate markers in each cluster of differentially expressed genes between the two conditions. We focused our analysis on Pseudomonas aeruginosa due to high urogenital virulence, and immunomodulation capabilities by pattern recognition receptors and inflammasome.
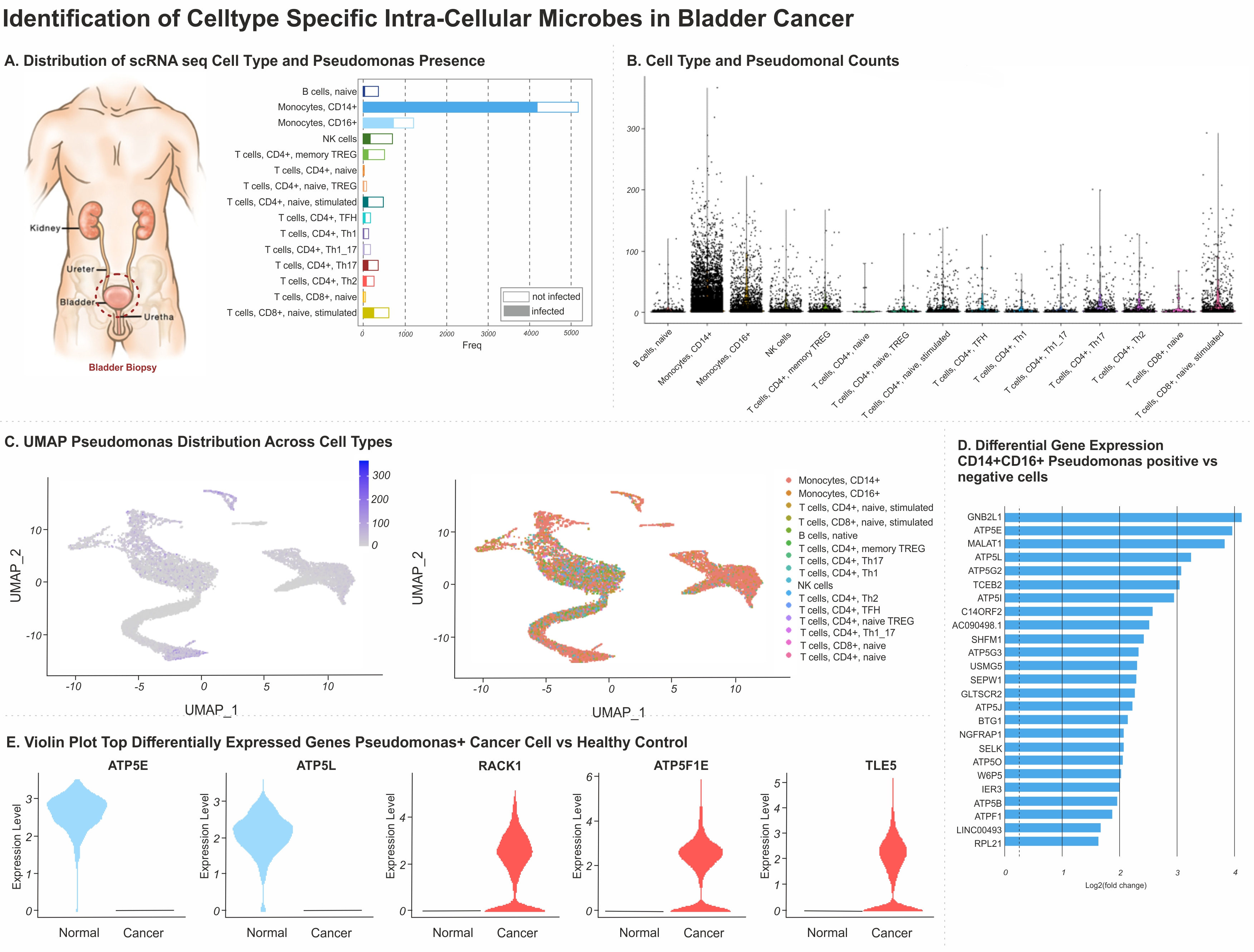
Results: Patients with history of high-grade BC demonstrated increased presence of Aeromonas (175,473 reads, 54%), Pseudomonas (75,002 reads, 26.73%), and Bacillus (15,410 reads, 5.49%) in their tissue samples, not visualized in healthy controls (counts <10). Among 8,324 cells (across 7 patients), classical CD14+ monocyte subtype was most likely to be exposed to Pseudomonas species (4150 /5069, 81.8%), followed by non-classical CD16+ monocytes (722/1040, 69.4%), and CD8+ Tcells (348/680, 51.1%). (Figure 1A-C). Relative gene expression level between exposed and non-exposed monocytes demonstrated total 25 upregulated genes, with increased expression GNBL21 (aka: RACK1, log10 4.1), MALAT1(log10 3.7), TCEB2(log10 2.9) (increasing proliferation, migration, and reduction of cell autophagy) compared to non-infected, and normal controls. Pseudomonas involvement of non-monocyte cells (CD8+, CD4+Th17, CD4+ naïve) was associated with disease progression (2/5 patients), as well as increased expression of IFI6/27(interferon alpha inducible protein 6 &27), ITAC (interferon inducible T cell alpha-chemoattractant).
Conclusions: Altogether, our analysis depicted the distribution of intracellular bacteria not previously recognized within tumor microenvironment utilizing scRNAseq, and revealed overexpression of RACK1 in Pseudomonas positive cells, associated with apoptosis resistance. SOURCE OF
Funding: Bladder Cancer Advocacy Network Young Investigator Award
Podium Session
Session: PD25: Bladder Cancer: Basic Research & Pathophysiology II
PD25-12: Identification of intratumor pathogens from single-cell RNA sequencing, cellular dysfunction and immune response
Sunday, April 30, 2023
8:50 AM – 9:00 AM CST
Location: S404A
- LB
Podium Presenter(s)
Introduction: Pathogenic microbes can induce cellular dysfunction and alters immune response to therapy. Taking advantage of single-cell RNA-sequencing (scRNA-seq), we aim to assess the transcriptomic features of the single cell level to identify intracellular pathogens in patients with high grade bladder cancer.
Methods: CellRanger and Alevin were utilized to extract cell barcodes and unique molecular identifiers. Low complexity reads <20 was considered negative, and PCR duplicates were removed. Seuret Findmarkers, scIntegrate, and scDensity Plot were utilized to normalize the data, cluster the cells, and calculate markers in each cluster of differentially expressed genes between the two conditions. We focused our analysis on Pseudomonas aeruginosa due to high urogenital virulence, and immunomodulation capabilities by pattern recognition receptors and inflammasome.
Results: Patients with history of high-grade BC demonstrated increased presence of Aeromonas (175,473 reads, 54%), Pseudomonas (75,002 reads, 26.73%), and Bacillus (15,410 reads, 5.49%) in their tissue samples, not visualized in healthy controls (counts <10). Among 8,324 cells (across 7 patients), classical CD14+ monocyte subtype was most likely to be exposed to Pseudomonas species (4150 /5069, 81.8%), followed by non-classical CD16+ monocytes (722/1040, 69.4%), and CD8+ Tcells (348/680, 51.1%). (Figure 1A-C). Relative gene expression level between exposed and non-exposed monocytes demonstrated total 25 upregulated genes, with increased expression GNBL21 (aka: RACK1, log10 4.1), MALAT1(log10 3.7), TCEB2(log10 2.9) (increasing proliferation, migration, and reduction of cell autophagy) compared to non-infected, and normal controls. Pseudomonas involvement of non-monocyte cells (CD8+, CD4+Th17, CD4+ naïve) was associated with disease progression (2/5 patients), as well as increased expression of IFI6/27(interferon alpha inducible protein 6 &27), ITAC (interferon inducible T cell alpha-chemoattractant).
Conclusions: Altogether, our analysis depicted the distribution of intracellular bacteria not previously recognized within tumor microenvironment utilizing scRNAseq, and revealed overexpression of RACK1 in Pseudomonas positive cells, associated with apoptosis resistance. SOURCE OF
Funding: Bladder Cancer Advocacy Network Young Investigator Award

