Back
Introduction: Repeat transurethral resection (re-TUR) in non-muscle invasive bladder cancer (NMIBC) is recommended for patients with high-risk Ta and T1 lesions, and often more extensively samples the tumor base and detrusor musculature. In this hypothesis-generating analysis, we seek to characterize the variability in genomic alterations between index and re-TUR specimens as they may be implicated in muscle-invasion as well as choice of and response to intravesical therapies.
Methods: Baseline mutational profiling of six index NMIBC tumors and paired repeat resections was performed via PredicineWES+™ whole-exome sequencing across 20,000 genes with boosted sequencing of 600 cancer-related genes in patients with high-risk Ta and T1 lesions on index resection. Non-synonymous (NS) mutations observed in index and re-TUR specimens identified in a minimum of two samples are reported.
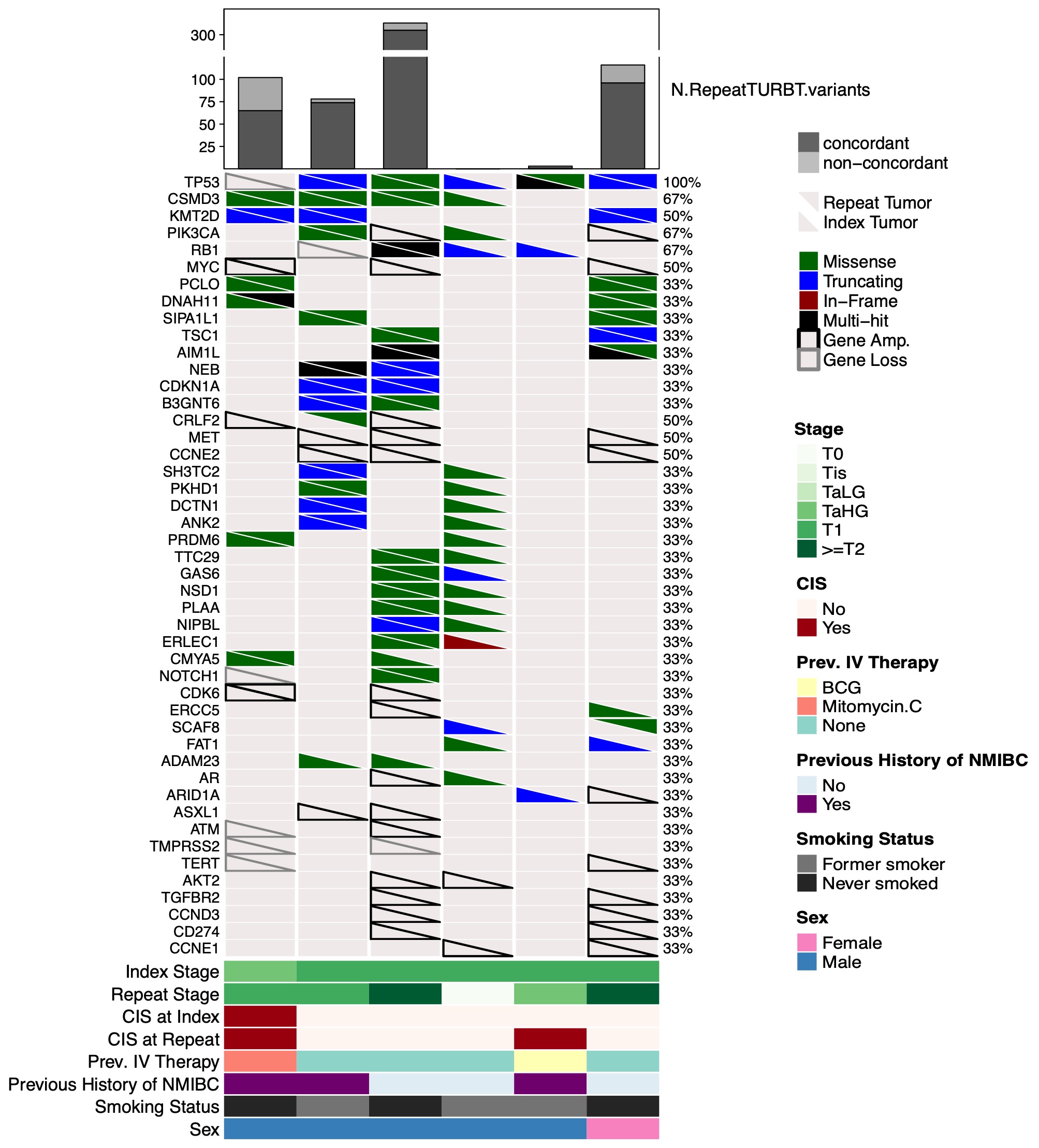
Results: Five and one patients had T1HG and TaHG index TURBT, respectively and two patients were upstaged to pT2 on re-TUR. As shown in the OncoPrint (Figure 1), 46 genes with NS mutations in at least two samples were identified including TP53, PIK3CA, and ARID1A. A median of 140 (range 48-420) and 90 NS (range 0-313) mutations were identified in the index and re-TUR specimens, respectively. On average, in patients with malignancy on re-TUR, lesions were 83% concordant with the index (range 33%-97%). One patient was pT0 on re-TUR for which no concordant variants were identified. Of non-concordant mutations, cell-cycle (CCND3, CCNE1, CCNE2) and PD-L1 (CD274) gene amplifications were uniquely expressed on the index lesion.
Conclusions: Genomic alterations between index and re-TUR lesions are highly concordant even when pathologic upstaging occurs. However, index lesions harbored unique mutations in cell cycle and immune checkpoint genes. Surface tumor cells may behave differently than base tumor cells, with varying therapeutic targets. SOURCE OF
Funding: None
Moderated Poster Session
Session: MP14: Bladder Cancer: Basic Research & Pathophysiology I
MP14-08: Characterizing Variability in Genomic Alterations between Index and Repeat Transurethral Resection Lesions in Non-Muscle Invasive Bladder Cancer
Friday, April 28, 2023
1:00 PM – 3:00 PM CST
Location: S403

Prithvi Murthy, MD
Fellow in Urologic Oncology
Moffitt Cancer Center
Poster Presenter(s)
Introduction: Repeat transurethral resection (re-TUR) in non-muscle invasive bladder cancer (NMIBC) is recommended for patients with high-risk Ta and T1 lesions, and often more extensively samples the tumor base and detrusor musculature. In this hypothesis-generating analysis, we seek to characterize the variability in genomic alterations between index and re-TUR specimens as they may be implicated in muscle-invasion as well as choice of and response to intravesical therapies.
Methods: Baseline mutational profiling of six index NMIBC tumors and paired repeat resections was performed via PredicineWES+™ whole-exome sequencing across 20,000 genes with boosted sequencing of 600 cancer-related genes in patients with high-risk Ta and T1 lesions on index resection. Non-synonymous (NS) mutations observed in index and re-TUR specimens identified in a minimum of two samples are reported.
Results: Five and one patients had T1HG and TaHG index TURBT, respectively and two patients were upstaged to pT2 on re-TUR. As shown in the OncoPrint (Figure 1), 46 genes with NS mutations in at least two samples were identified including TP53, PIK3CA, and ARID1A. A median of 140 (range 48-420) and 90 NS (range 0-313) mutations were identified in the index and re-TUR specimens, respectively. On average, in patients with malignancy on re-TUR, lesions were 83% concordant with the index (range 33%-97%). One patient was pT0 on re-TUR for which no concordant variants were identified. Of non-concordant mutations, cell-cycle (CCND3, CCNE1, CCNE2) and PD-L1 (CD274) gene amplifications were uniquely expressed on the index lesion.
Conclusions: Genomic alterations between index and re-TUR lesions are highly concordant even when pathologic upstaging occurs. However, index lesions harbored unique mutations in cell cycle and immune checkpoint genes. Surface tumor cells may behave differently than base tumor cells, with varying therapeutic targets. SOURCE OF
Funding: None

