Back
Introduction: Novel predictive tools to guide treatment decision are required and novel therapies are needed. Immunotherapies are under investigation and hold strong promise, however efficacy is still limited. On this line, a comprehensive analysis of the immune infiltrate in BC would reveal mechanisms of resistance and unveil novel targets.
Methods: Here we investigated the profile of the immune microenvironment in NMBC and MIBC patients by high dimensional single cell analysis, including scRNAseq and Imaging Mass Cytometry. For the scRNAseq analysis, we enrolled NMBC and MIBC patients and we FACS sorted CD45+ immune cells and CD45- epithelia and stromal cells. Cells were then processed by 3’-based scRNA-seq strategy (10X Genomics) that revealed the transcriptional landscape of sorted populations. Additional patients were enrolled for the Cytomass analysis. Tissue sections were snap frozen, stained and acquired by Hyperion system. The primary endpoint was to unveil differences in the composition and tumor and immune cells in NMIBC and MIBC tumors.
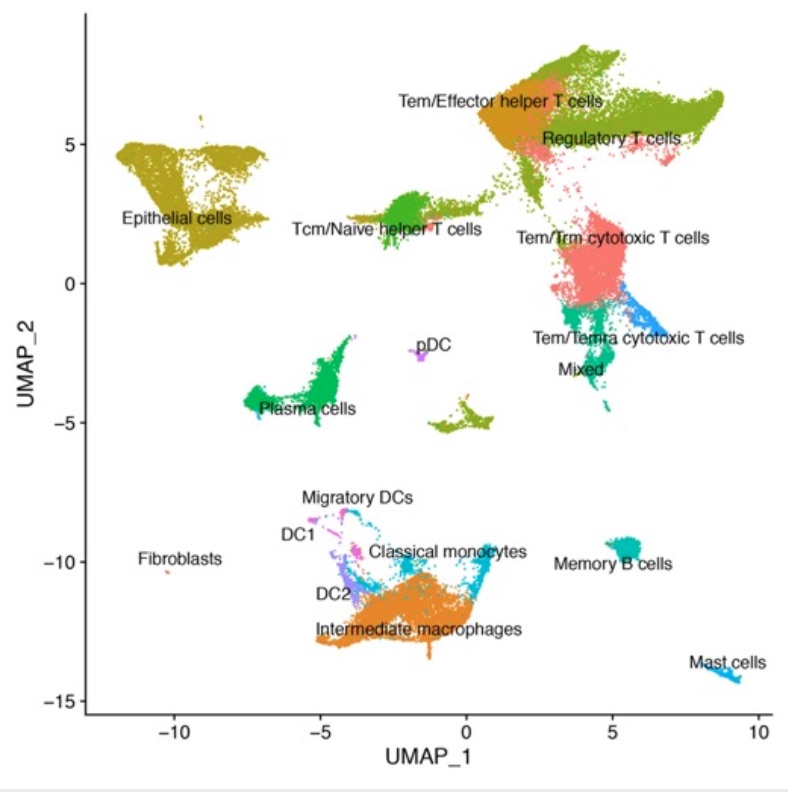
Results: The entire cohort consisted of 14 patients with clinically significant bladder cancer who underwent radical cystectomy and TURBT. Our analysis identified differences in immune cells abundance and activation state when comparing NMBC and MIBC samples. Modulations were particularly evident for a subset of PLIN2+ Tumor associated macrophages (TAMs) that was enriched in invasive tumors (Fold change= 8.37). In the lymphoid compartment, T regulatory cells (Tregs) showed strong transcriptional changes between NMBC and MIBC samples, that suggest a higher fitness and suppressive function in Tregs that infiltrate MIBC. In addition, our analysis revealed a high heterogeneity in the composition of tumor cells in both cohorts (Fig. 1). Finally, Cytomass analysis unveiled modulations in immune cells activation and spatial localization when comparing NMBC and MIBC that will be further investigated
Conclusions: Our study supports the effectiveness of single cell RNA sequencing in the dissection of the immune landscape in BC and identified immune changes when comparing neoplastic tissue from MIBC with NMBC patients. SOURCE OF
Funding: None
Moderated Poster Session
Session: MP14: Bladder Cancer: Basic Research & Pathophysiology I
MP14-11: Single cell-based immune profiling of the tumor and its immune microenvironment revealed differences between NMIBC and MIBC: implication for immuno-therapy
Friday, April 28, 2023
1:00 PM – 3:00 PM CST
Location: S403
Poster Presenter(s)
Introduction: Novel predictive tools to guide treatment decision are required and novel therapies are needed. Immunotherapies are under investigation and hold strong promise, however efficacy is still limited. On this line, a comprehensive analysis of the immune infiltrate in BC would reveal mechanisms of resistance and unveil novel targets.
Methods: Here we investigated the profile of the immune microenvironment in NMBC and MIBC patients by high dimensional single cell analysis, including scRNAseq and Imaging Mass Cytometry. For the scRNAseq analysis, we enrolled NMBC and MIBC patients and we FACS sorted CD45+ immune cells and CD45- epithelia and stromal cells. Cells were then processed by 3’-based scRNA-seq strategy (10X Genomics) that revealed the transcriptional landscape of sorted populations. Additional patients were enrolled for the Cytomass analysis. Tissue sections were snap frozen, stained and acquired by Hyperion system. The primary endpoint was to unveil differences in the composition and tumor and immune cells in NMIBC and MIBC tumors.
Results: The entire cohort consisted of 14 patients with clinically significant bladder cancer who underwent radical cystectomy and TURBT. Our analysis identified differences in immune cells abundance and activation state when comparing NMBC and MIBC samples. Modulations were particularly evident for a subset of PLIN2+ Tumor associated macrophages (TAMs) that was enriched in invasive tumors (Fold change= 8.37). In the lymphoid compartment, T regulatory cells (Tregs) showed strong transcriptional changes between NMBC and MIBC samples, that suggest a higher fitness and suppressive function in Tregs that infiltrate MIBC. In addition, our analysis revealed a high heterogeneity in the composition of tumor cells in both cohorts (Fig. 1). Finally, Cytomass analysis unveiled modulations in immune cells activation and spatial localization when comparing NMBC and MIBC that will be further investigated
Conclusions: Our study supports the effectiveness of single cell RNA sequencing in the dissection of the immune landscape in BC and identified immune changes when comparing neoplastic tissue from MIBC with NMBC patients. SOURCE OF
Funding: None


