Back
Introduction: Among patients with AJCC Stage IIC+ cancer, a subset of patients are genomically at highest risk of progression with very high Decipher =0.95 scores. This group of patients is distinct and not present with AJCC I-IIB prostate cancer. As genomic changes may predict potential benefit from molecular based therapies, we sought to identify incidence of genomic alterations in this population.
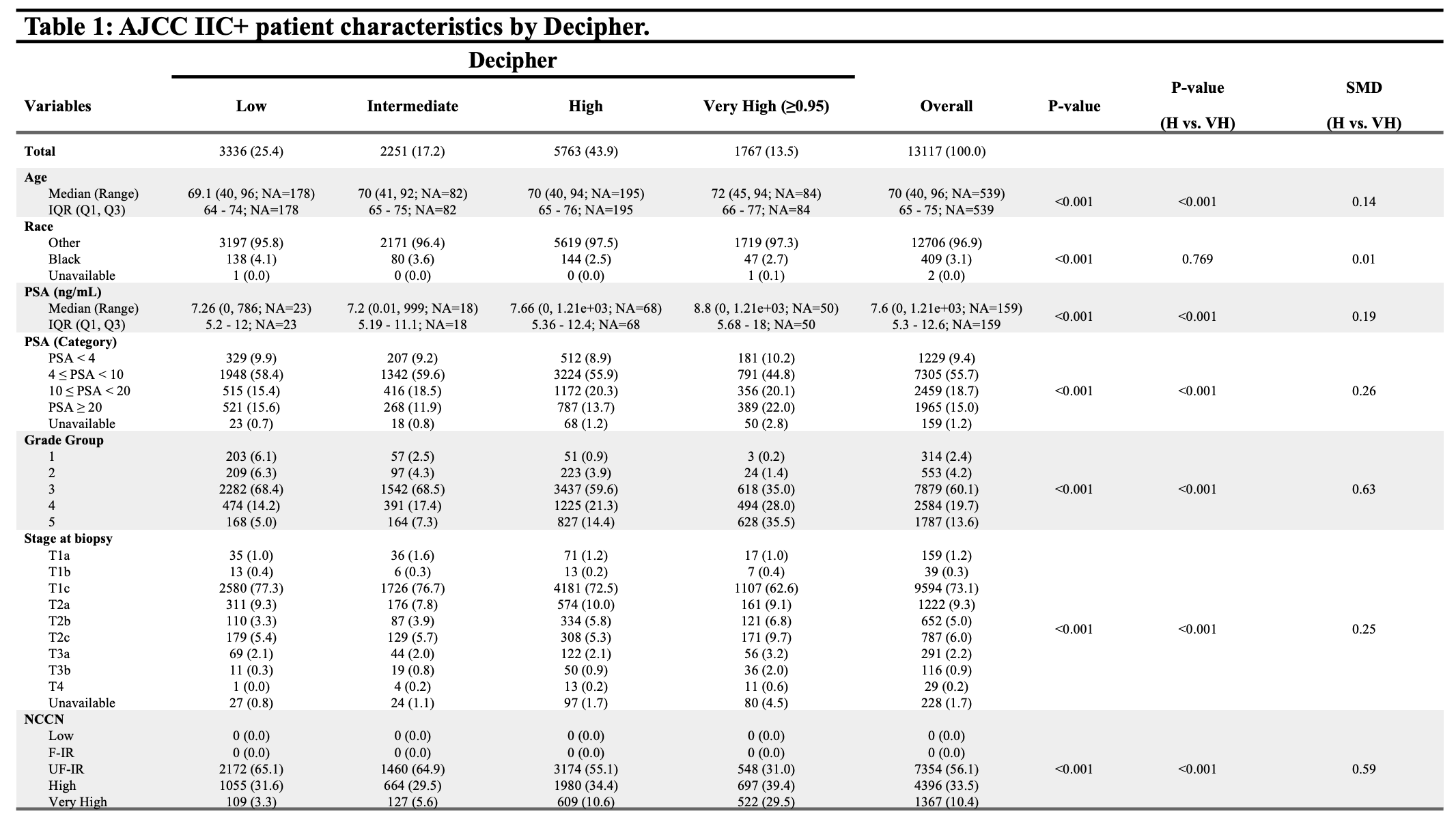
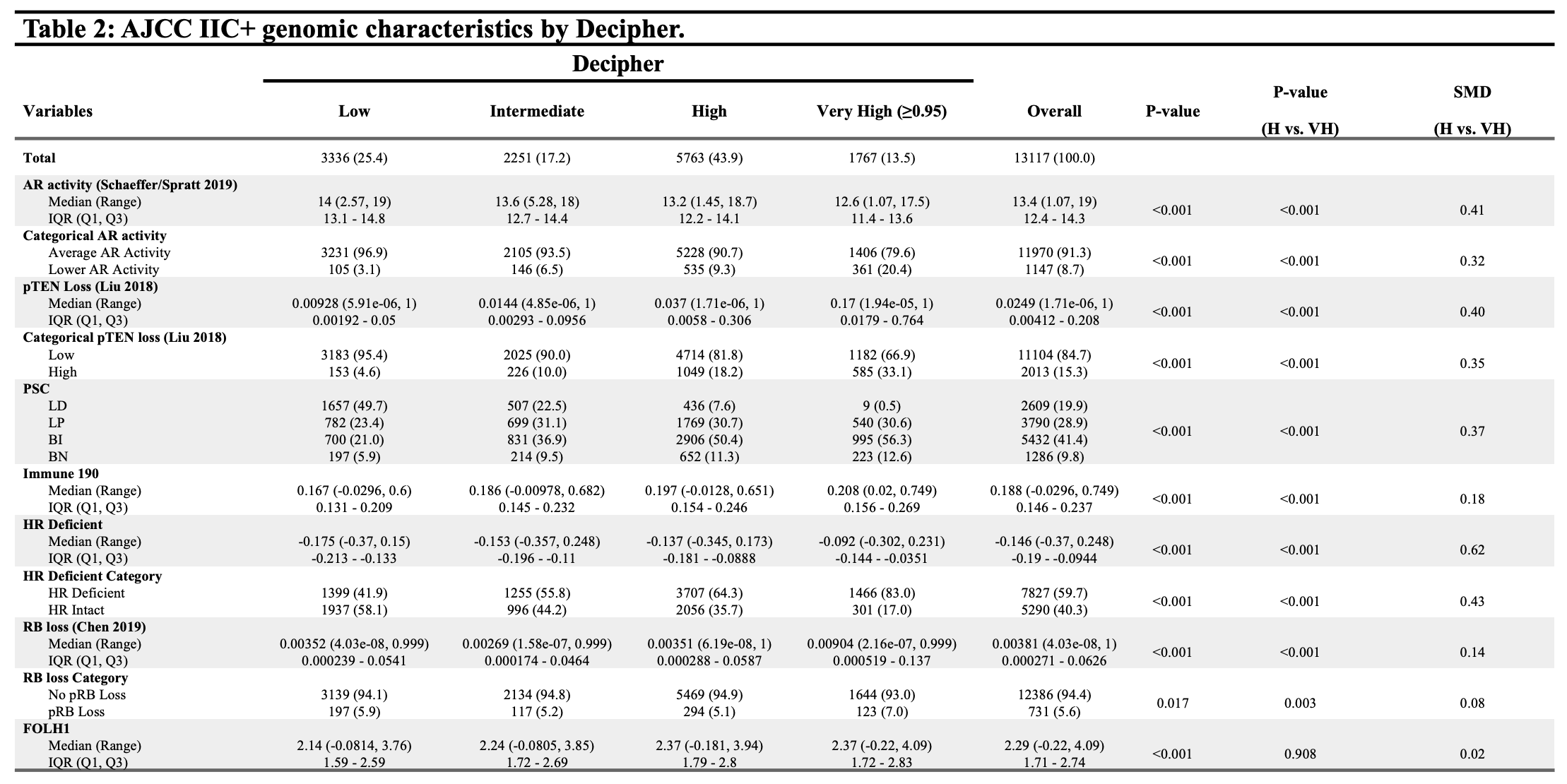
Methods: We analyzed the transcriptomes of 13,117 men diagnosed with AJCC stage IIC+ localized prostate cancer. We compared signature readouts for androgen receptor activity (AR-A), PTEN loss, homologous repair deficiency, Rb loss, immune activity, and PSMA (FOLH1) expression, as well as prostate subtyping classifier (PSC) based on cell of origin and molecular pathways. Standardized mean differences (SMD) were calculated to compare size effect, with significant effect size defined as 0.2.
Results: 1767 men with AJCC Stage IIC+ cancer were found to have Decipher score =0.95. Patients with Decipher =0.95 possessed lower AR activity, as well as higher rates of PTEN loss and HR deficiency compared to men with high Decipher 0.6-0.95 (SMD 0.32, 0.40, and 0.43). There was a lower incidence of luminal differentiated (LD), and higher incidence of basal immune (BI) and basal neuroendocrine (BN) subtypes among those with Decipher =0.95. Rates of Rb loss remained low across all Decipher subgroups, and there was no significant difference in FOLH1 expression comparing men with Decipher 0.6-0.95 and Decipher =0.95.
Conclusions: Patients with very high Decipher =0.95 localized prostate cancer have a distinct molecular profile, and may benefit from PARP inhibitors. Further investigation is required to understand how these molecular signatures may drive progression and metastasis in patients who are identified as very high risk on genomic classifier testing. SOURCE OF
Funding: None
Moderated Poster Session
Session: MP17: Prostate Cancer: Markers
MP17-01: Differential Transcriptomic Indicators of Potential Therapeutic Response to Targeted Therapy for AJCC IIC+ Localized Prostate Cancer with Decipher ≥0.95
Friday, April 28, 2023
1:00 PM – 3:00 PM CST
Location: S504

Eric V. Li, MD (he/him/his)
Northwestern University Feinberg School of Medicine
Poster Presenter(s)
Introduction: Among patients with AJCC Stage IIC+ cancer, a subset of patients are genomically at highest risk of progression with very high Decipher =0.95 scores. This group of patients is distinct and not present with AJCC I-IIB prostate cancer. As genomic changes may predict potential benefit from molecular based therapies, we sought to identify incidence of genomic alterations in this population.
Methods: We analyzed the transcriptomes of 13,117 men diagnosed with AJCC stage IIC+ localized prostate cancer. We compared signature readouts for androgen receptor activity (AR-A), PTEN loss, homologous repair deficiency, Rb loss, immune activity, and PSMA (FOLH1) expression, as well as prostate subtyping classifier (PSC) based on cell of origin and molecular pathways. Standardized mean differences (SMD) were calculated to compare size effect, with significant effect size defined as 0.2.
Results: 1767 men with AJCC Stage IIC+ cancer were found to have Decipher score =0.95. Patients with Decipher =0.95 possessed lower AR activity, as well as higher rates of PTEN loss and HR deficiency compared to men with high Decipher 0.6-0.95 (SMD 0.32, 0.40, and 0.43). There was a lower incidence of luminal differentiated (LD), and higher incidence of basal immune (BI) and basal neuroendocrine (BN) subtypes among those with Decipher =0.95. Rates of Rb loss remained low across all Decipher subgroups, and there was no significant difference in FOLH1 expression comparing men with Decipher 0.6-0.95 and Decipher =0.95.
Conclusions: Patients with very high Decipher =0.95 localized prostate cancer have a distinct molecular profile, and may benefit from PARP inhibitors. Further investigation is required to understand how these molecular signatures may drive progression and metastasis in patients who are identified as very high risk on genomic classifier testing. SOURCE OF
Funding: None


