Back
Introduction: Detection of a microbiome profile coupled with advanced genetic sequencing has opened a new research frontier. We hypothesized that the semen microbiome profile in men with non-obstructive azoospermia (NOA) would differ from that of fertile controls (FC).
Methods: NOA was diagnosed using semen analysis and clinical phenotype (small testis volume, FSH >10 IU/mL). FC were identified as men undergoing vasectomy after fathering at least one child. Semen samples were collected and underwent Next Generation Sequencing (NGS) including quantitative PCR and 16s rRNA V1/V2 region comprehensive sequencing using Illumina MiSeq technology. Data was processes and analyzed in dada2. Alpha diversity was analyzed via Margalef. Beta diversity was analyzed via Bray-Curtis difference. Differential abundance was evaluated with ANCOMBC procedure.
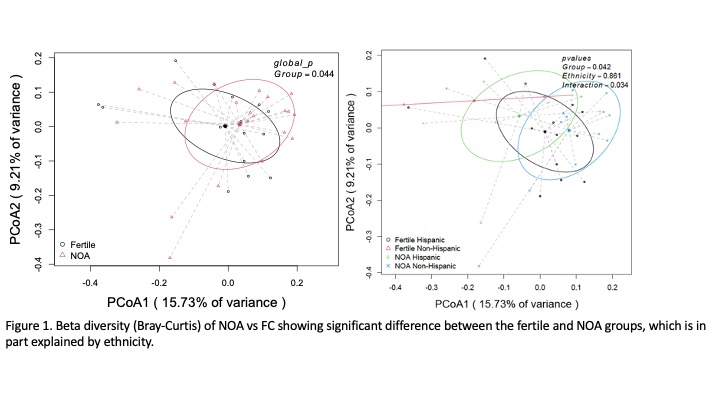
Results: After rarefaction analysis, 33 samples were included (18 NOA and 15 FC) determined samples had sufficient sequencing depth to capture microbiome diversity. Alpha diversity was lower in the NOA men but did not reach statistical significance. Beta diversity showed a difference between the NOA and fertile groups primarily driven by differences in ethnicity. At the phylum level, NOA patients demonstrated lower levels of Proteobacteria and Firmicutes and higher levels of Actinobacteriota compared to the FC group consistent with prior published work. At the genus level, the known uropathogens Ureaplasma (1.3% vs 0.16%) and Eschrichia/Shigella (2.26% vs 0.42%) were higher in the NOA group compared to fertile controls. Other notable differences were Raoultella (39-fold higher in Fertile – 2% vs 0.09%) and Sneathia (9.5-fold higher in Fertile – 1% vs. 0.1%). Differential abundance analysis largely confirmed these findings.
Conclusions: Compared to fertile controls, men with NOA have a less diverse seminal microbiome enriched for known pathogens and several understudied organisms. This work supports the presence of a testicular microbiome, which may become dysbiotic with infertility and could be leveraged as a biomarker to guide treatment decision making in the future. SOURCE OF
Funding: MicrogenDX
Moderated Poster Session
Session: MP01: Infertility: Basic Research & Pathophysiology
MP01-07: A Next-Generation Sequencing Analysis of the Semen Microbiome in Men with Non-Obstructive Azoospermia
Friday, April 28, 2023
7:00 AM – 9:00 AM CST
Location: S401A

Katherine Campbell, BS (she/her/hers)
Desai Sethi Urology Institute
Poster Presenter(s)
Introduction: Detection of a microbiome profile coupled with advanced genetic sequencing has opened a new research frontier. We hypothesized that the semen microbiome profile in men with non-obstructive azoospermia (NOA) would differ from that of fertile controls (FC).
Methods: NOA was diagnosed using semen analysis and clinical phenotype (small testis volume, FSH >10 IU/mL). FC were identified as men undergoing vasectomy after fathering at least one child. Semen samples were collected and underwent Next Generation Sequencing (NGS) including quantitative PCR and 16s rRNA V1/V2 region comprehensive sequencing using Illumina MiSeq technology. Data was processes and analyzed in dada2. Alpha diversity was analyzed via Margalef. Beta diversity was analyzed via Bray-Curtis difference. Differential abundance was evaluated with ANCOMBC procedure.
Results: After rarefaction analysis, 33 samples were included (18 NOA and 15 FC) determined samples had sufficient sequencing depth to capture microbiome diversity. Alpha diversity was lower in the NOA men but did not reach statistical significance. Beta diversity showed a difference between the NOA and fertile groups primarily driven by differences in ethnicity. At the phylum level, NOA patients demonstrated lower levels of Proteobacteria and Firmicutes and higher levels of Actinobacteriota compared to the FC group consistent with prior published work. At the genus level, the known uropathogens Ureaplasma (1.3% vs 0.16%) and Eschrichia/Shigella (2.26% vs 0.42%) were higher in the NOA group compared to fertile controls. Other notable differences were Raoultella (39-fold higher in Fertile – 2% vs 0.09%) and Sneathia (9.5-fold higher in Fertile – 1% vs. 0.1%). Differential abundance analysis largely confirmed these findings.
Conclusions: Compared to fertile controls, men with NOA have a less diverse seminal microbiome enriched for known pathogens and several understudied organisms. This work supports the presence of a testicular microbiome, which may become dysbiotic with infertility and could be leveraged as a biomarker to guide treatment decision making in the future. SOURCE OF
Funding: MicrogenDX

