Back
Purpose: Historically, immunoassay technologies, reagents, and labeling techniques were systemically variable and required duplicate-well analysis to ensure accurate and precise analytical results. This led to duplicate-well analysis becoming a standard practice in pharmacokinetic, quantitative biomarker, and immunogenicity immunoassays. Of note, duplicate-well analyses are commonly used in immunogenicity immunoassays to construct statistical distributions to calculate assay cut points (CP) and for overall acceptance and rejection of plates. However, duplicate-well analysis has increasingly become unnecessary due to the advances in immunoassay technologies and critical reagent generation leading to improvement in overall assay precision and robustness and the possibility of moving to single-well analysis without a reduction in data quality and integrity.
Purpose: To evaluate the use of single-well analysis over duplicate-well analysis in less variable populations, such as preclinical populations.
Methods: In a case study, we used regression analysis to evaluate the distribution of percent inhibition and signal to noise (S/N) ratios to determine if there is any significant difference in the distributions whether we use duplicate 1, duplicate 2, or the duplicate mean values. To look for insignificant differences, we used α = 0.10 as a stricter significance threshold to be met. To compare the mean of the distribution for duplicate 1, duplicate 2, and duplicate mean values, we used ordinary least squares (OLS) regression. We used quantile regression to compare the 25th, 50th, and 75th percentiles of the distributions using duplicate 1, duplicate 2, and duplicate mean values. Additionally, separate screening and confirmatory CP analyses were performed on the duplicate 1, duplicate 2, and duplicate mean data sets and were evaluated independently using the corresponding CP value for tier 1 and tier 2. Lastly, a comparison of positive control classification and the experimental results of assay characterization experiments, e.g., drug and target tolerance, matrix interference, selectivity, sensitivity, and stability, was performed to determine differences in experimental conclusions (pass vs fail) based on whether duplicate 1, duplicate 2, or duplicate mean data was used.
Results: We observed no significant difference in the mean of the distribution whether we use duplicate 1, duplicate 2, or the duplicate mean values. The OLS regression resulted in p-values ranging from 0.914 - 0.996 in rat and 0.738 - 0.814 in non-human primates (NHP) thereby implying that no means are significantly different from another. The quantile regression to compare the 25th, 50th, and 75th percentiles of the distributions using duplicate 1, duplicate 2, and duplicate mean values yielded p-values ranging from 0.262 - 0.944 in rat and from 0.201 - 0.944 in NHP, implying that there is no percentile in one dataset that is significantly different from another. For the CP analyses performed on the duplicate 1, duplicate 2, and duplicate mean data sets, we observed screening CP values of 1.16, 1.19 and 1.19 (S/N), and confirmatory CP values of 25.0%, 29.1%, and 27.3% inhibition, respectively. These differences between the CP values for tiers I and II were confirmed to be not statistically significant (p-values ranged from 0.414 - 1.00). And finally, the comparison of positive control classification and the experimental results of assay characterization experiments, resulted in no difference in experimental conclusions (pass vs fail) whether duplicate 1, duplicate 2, or duplicate mean data was used.
Conclusion: Through this case study, we are able to demonstrate that similar results to duplicate-well analysis are obtained by using single-well analysis in less variable populations, such as preclinical populations.
References: Ye, Z., Tu, J., Midde, K., Edwards, M., & Bennett, P. (2018). Singlicate analysis: Should this be the default for biomarker measurements using ligand-binding assays? Bioanalysis, 10(12), 909–912. https://doi.org/10.4155/bio-2018-0067
Barfield, M., Goodman, J., Hood, J., & Timmerman, P. (2020). European Bioanalysis Forum recommendation on SINGLICATE analysis for ligand binding assays: Time for a new mindset. Bioanalysis, 12(5), 273–284. https://doi.org/10.4155/bio-2019-0298
Acknowledgments: Disclosure: Katelyn Crizer, PhD was employed at Altavant Sciences, Inc. during the length of this study. Dr. Crizer is currently employed at BioData Solutions, LLC (the consulting organization on this project).
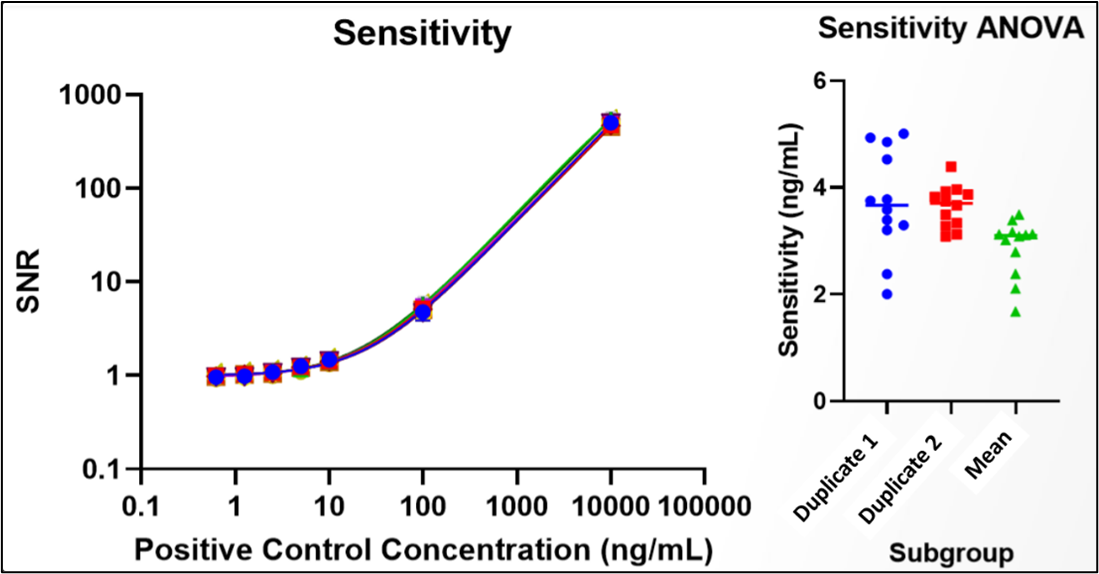
Sensitivity determination for Screening Cut Point
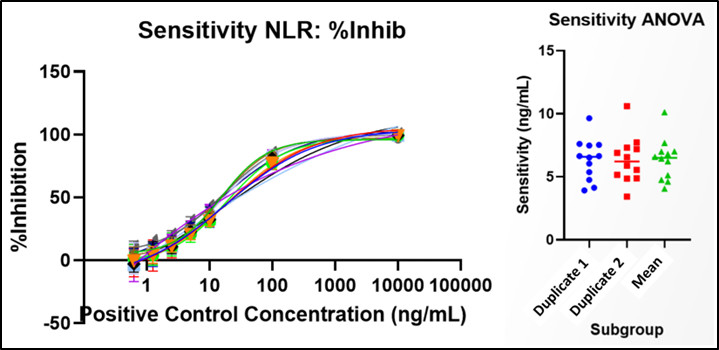
Sensitivity determination for Confirmatory Cut Point
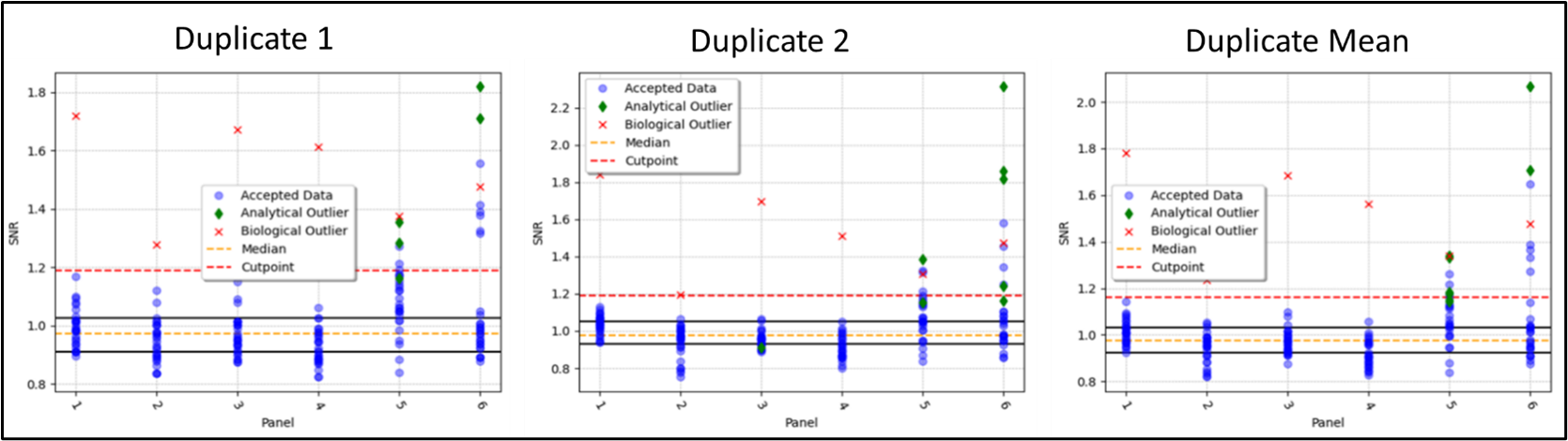
Scatter plot for Screening Cut Point
Bioanalytics - Biomolecular - Immunogenicity
Category: Late Breaking Poster Abstract
(W1030-09-49) An Evaluation of the Effect of Duplicate- and Single- Well Analyses on Assay Cut Point Calculations and Characterization Experiments in Preclinical Immunogenicity Assays
Wednesday, October 19, 2022
10:30 AM – 11:30 AM ET
- KC
Katelyn Crizer, Ph.D.
BioData Solutions LLC
Lawrence, Kansas, United States - JM
Janine Micheli, Ph.D.
PPD Laboratories
Richmond, Virginia, United States
Main Author(s)
Presenting Author(s)
Purpose: Historically, immunoassay technologies, reagents, and labeling techniques were systemically variable and required duplicate-well analysis to ensure accurate and precise analytical results. This led to duplicate-well analysis becoming a standard practice in pharmacokinetic, quantitative biomarker, and immunogenicity immunoassays. Of note, duplicate-well analyses are commonly used in immunogenicity immunoassays to construct statistical distributions to calculate assay cut points (CP) and for overall acceptance and rejection of plates. However, duplicate-well analysis has increasingly become unnecessary due to the advances in immunoassay technologies and critical reagent generation leading to improvement in overall assay precision and robustness and the possibility of moving to single-well analysis without a reduction in data quality and integrity.
Purpose: To evaluate the use of single-well analysis over duplicate-well analysis in less variable populations, such as preclinical populations.
Methods: In a case study, we used regression analysis to evaluate the distribution of percent inhibition and signal to noise (S/N) ratios to determine if there is any significant difference in the distributions whether we use duplicate 1, duplicate 2, or the duplicate mean values. To look for insignificant differences, we used α = 0.10 as a stricter significance threshold to be met. To compare the mean of the distribution for duplicate 1, duplicate 2, and duplicate mean values, we used ordinary least squares (OLS) regression. We used quantile regression to compare the 25th, 50th, and 75th percentiles of the distributions using duplicate 1, duplicate 2, and duplicate mean values. Additionally, separate screening and confirmatory CP analyses were performed on the duplicate 1, duplicate 2, and duplicate mean data sets and were evaluated independently using the corresponding CP value for tier 1 and tier 2. Lastly, a comparison of positive control classification and the experimental results of assay characterization experiments, e.g., drug and target tolerance, matrix interference, selectivity, sensitivity, and stability, was performed to determine differences in experimental conclusions (pass vs fail) based on whether duplicate 1, duplicate 2, or duplicate mean data was used.
Results: We observed no significant difference in the mean of the distribution whether we use duplicate 1, duplicate 2, or the duplicate mean values. The OLS regression resulted in p-values ranging from 0.914 - 0.996 in rat and 0.738 - 0.814 in non-human primates (NHP) thereby implying that no means are significantly different from another. The quantile regression to compare the 25th, 50th, and 75th percentiles of the distributions using duplicate 1, duplicate 2, and duplicate mean values yielded p-values ranging from 0.262 - 0.944 in rat and from 0.201 - 0.944 in NHP, implying that there is no percentile in one dataset that is significantly different from another. For the CP analyses performed on the duplicate 1, duplicate 2, and duplicate mean data sets, we observed screening CP values of 1.16, 1.19 and 1.19 (S/N), and confirmatory CP values of 25.0%, 29.1%, and 27.3% inhibition, respectively. These differences between the CP values for tiers I and II were confirmed to be not statistically significant (p-values ranged from 0.414 - 1.00). And finally, the comparison of positive control classification and the experimental results of assay characterization experiments, resulted in no difference in experimental conclusions (pass vs fail) whether duplicate 1, duplicate 2, or duplicate mean data was used.
Conclusion: Through this case study, we are able to demonstrate that similar results to duplicate-well analysis are obtained by using single-well analysis in less variable populations, such as preclinical populations.
References: Ye, Z., Tu, J., Midde, K., Edwards, M., & Bennett, P. (2018). Singlicate analysis: Should this be the default for biomarker measurements using ligand-binding assays? Bioanalysis, 10(12), 909–912. https://doi.org/10.4155/bio-2018-0067
Barfield, M., Goodman, J., Hood, J., & Timmerman, P. (2020). European Bioanalysis Forum recommendation on SINGLICATE analysis for ligand binding assays: Time for a new mindset. Bioanalysis, 12(5), 273–284. https://doi.org/10.4155/bio-2019-0298
Acknowledgments: Disclosure: Katelyn Crizer, PhD was employed at Altavant Sciences, Inc. during the length of this study. Dr. Crizer is currently employed at BioData Solutions, LLC (the consulting organization on this project).

Sensitivity determination for Screening Cut Point

Sensitivity determination for Confirmatory Cut Point

Scatter plot for Screening Cut Point
