Back
Purpose: Alzheimer’s Disease (AD) has a strong spatial-temporal component to its progression, where different brain regions are affected by amyloid-beta (Aβ) plaque deposition at varying time points. Standard imaging and analysis platforms can neglect these details, as they lack the ability to pair high-yield whole-brain imaging with region-specific quantitation. Furthermore, many Aβ analyses require homogenization of tissue, prohibiting secondary analysis. To address this gap, we have developed a novel high-throughput whole-brain imaging pipeline for pre-clinical AD models to quantitatively track plaque progression as a function of brain region across time while producing indexed tissue sections for secondary staining and analysis.
Methods: Aβ plaques in the well characterized 5XFAD mouse model of AD were labeled with Methoxy-X04, an Aβ plaque-specific compound, prior to transcardial perfusion and whole-brain excision. The brains were processed on the TissueCyte Serial Two-Photon Plus (STP2) imaging platform to produce fully aligned multi-channel volumetric datasets, yielding high resolution 3D models of each brain. Registration to the Allen Mouse Brain Common Coordinate Framework (CCFv3) and region-specific plaque analysis were conducted to determine plaque size, density, and total number per animal brain. The regional density of dystrophic neurites (Lamp1) and microglia (Iba1) were segmented and quantified through secondary analysis to evaluate the signal in high and low Aβ growth rate regions for select sections and remapped to the original 3D whole-brain dataset.
Results: The analysis of whole-brain plaque distribution in the 5XFAD mouse model revealed distinct spatial-temporal changes across the brains (Figure 1, Figure 2). STP2 imaging, combined with CCFv3 mapping and secondary analysis of Lamp1 and Iba1 allowed for targeted evaluation of Aβ plaques, and a comparison of regional molecular changes between models.
Conclusion: This novel technology has great promise for quantifying the spatial-temporal Aβ plaque efficacy of AD animal models, and the production of translatable pre-clinical AD drug discovery data. The high sensitivity and precision of the STP2 platform can benefit region-specific disease progression compared to standard laboratory approaches.
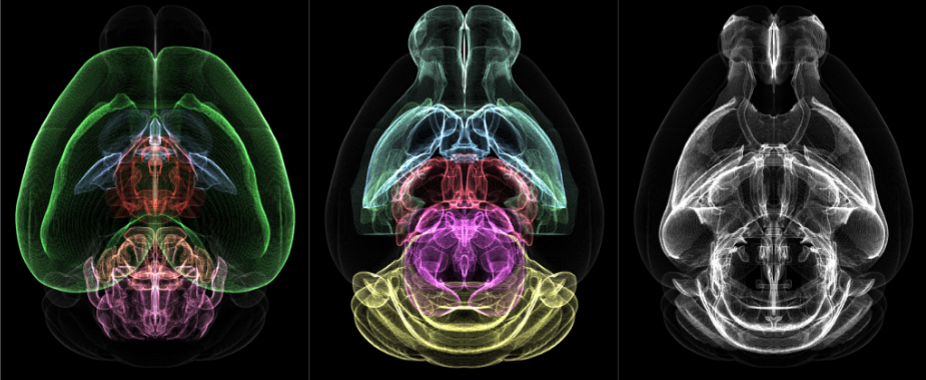
Figure 1. Volumetric profiles of the major brain regions (colorized) from the Allen Mouse Brain Common Coordinate Framework.
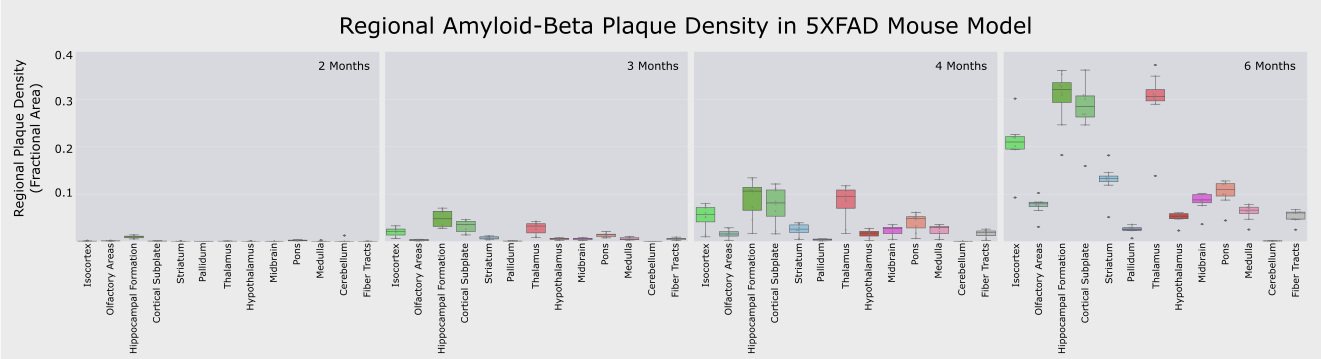
Figure 2. Plots of amyloid-beta plaque density for the major brain regions (colorized as in Figure 1) across the 4 time points (2,3,4 and 6 months) of 5XFAD female mice, demonstrating a significant density accumulation across disease progression (* p<0.05).
Preclinical Development - Chemical - Translation
Category: Late Breaking Poster Abstract
(T1330-02-07) A Novel Whole-Brain Imaging Pipeline for High Resolution Spatial and Temporal Measurements of Amyloid-Beta Plaque Dynamics in Pre-clinical Alzheimer’s Disease Animal Models
Tuesday, October 18, 2022
1:30 PM – 2:30 PM ET
- GF
Gianna Ferron
Scientist I
TissueVision Inc. - GF
Gianna Ferron
Scientist I
TissueVision Inc.
Presenting Author(s)
Main Author(s)
Purpose: Alzheimer’s Disease (AD) has a strong spatial-temporal component to its progression, where different brain regions are affected by amyloid-beta (Aβ) plaque deposition at varying time points. Standard imaging and analysis platforms can neglect these details, as they lack the ability to pair high-yield whole-brain imaging with region-specific quantitation. Furthermore, many Aβ analyses require homogenization of tissue, prohibiting secondary analysis. To address this gap, we have developed a novel high-throughput whole-brain imaging pipeline for pre-clinical AD models to quantitatively track plaque progression as a function of brain region across time while producing indexed tissue sections for secondary staining and analysis.
Methods: Aβ plaques in the well characterized 5XFAD mouse model of AD were labeled with Methoxy-X04, an Aβ plaque-specific compound, prior to transcardial perfusion and whole-brain excision. The brains were processed on the TissueCyte Serial Two-Photon Plus (STP2) imaging platform to produce fully aligned multi-channel volumetric datasets, yielding high resolution 3D models of each brain. Registration to the Allen Mouse Brain Common Coordinate Framework (CCFv3) and region-specific plaque analysis were conducted to determine plaque size, density, and total number per animal brain. The regional density of dystrophic neurites (Lamp1) and microglia (Iba1) were segmented and quantified through secondary analysis to evaluate the signal in high and low Aβ growth rate regions for select sections and remapped to the original 3D whole-brain dataset.
Results: The analysis of whole-brain plaque distribution in the 5XFAD mouse model revealed distinct spatial-temporal changes across the brains (Figure 1, Figure 2). STP2 imaging, combined with CCFv3 mapping and secondary analysis of Lamp1 and Iba1 allowed for targeted evaluation of Aβ plaques, and a comparison of regional molecular changes between models.
Conclusion: This novel technology has great promise for quantifying the spatial-temporal Aβ plaque efficacy of AD animal models, and the production of translatable pre-clinical AD drug discovery data. The high sensitivity and precision of the STP2 platform can benefit region-specific disease progression compared to standard laboratory approaches.

Figure 1. Volumetric profiles of the major brain regions (colorized) from the Allen Mouse Brain Common Coordinate Framework.

Figure 2. Plots of amyloid-beta plaque density for the major brain regions (colorized as in Figure 1) across the 4 time points (2,3,4 and 6 months) of 5XFAD female mice, demonstrating a significant density accumulation across disease progression (* p<0.05).
