Back
Purpose: We recently developed a prototype of an in vitro system, namely Emulator of SubCutaneous Absorption and Release (ESCAR), to emulate the in vivo subcutaneous (SC) environment. This study aimed to evaluate the ESCAR’s applicability in the assessment of biologics using a model protein bovine serum albumin (BSA).
Methods: ESCAR was fabricated by 3D printing technique with the aid of Computer-Aided Design (CAD). A DoE study was conducted to investigate the effects of fluid flow and hyaluronic acid (HA) concentration on drug (BSA) release. Molecular dynamics (MD) simulation was applied to study the electrostatic interaction between HA and BSA. The computational fluid dynamics (CFD) method was utilized to simulate the fluid flow inside ESCAR. Further, to reduce the experimental efforts during process/formulation change, a model migration method based on Bayesian inference was developed.
Results: The BSA release in HA solution was significantly slower than that in PBS, which was likely attributed to the following reasons: (1) lower viscosity in the medium; (2) higher resistance of BSA to diffusing through the characteristic length-scale of polymer chains; (3) electrostatic interaction between BSA and HA. Throughout the MD simulation, a total of 8 positively-charged residues (either arginine or lysine) in the BSA sequence were identified to have strong interactions with HA. Therefore, to reduce the interaction, scientists may consider mutating these residues to some neutral amino acids. Further, the fluid velocity and direction could be simulated by the CFD method. The results suggested that fluid flow/convection could substantially enhance the BSA release. To interpret this result from the in vivo perspective, the higher fluid flow/convection could lead to a faster migration from the SC injection site to the drainage site (e.g., lymphatic capillaries for most large molecules such as proteins). The DoE study pointed out that both fluid flow and HA concentration are critical parameters for the BSA release. The response surfaces based on the multivariate spline interpolation and Bagging method were developed to model the relationship between the input factors and the outputs (e.g., BSA release % at 2-hr, 4-hr, 8-hr, etc.). Further, a Bayesian interference-based method successfully migrated the model that was built in the old dataset (e.g., the dataset of 15-mg dose) to become a new model for the new dataset (e.g., the dataset of 30-mg dose). Compared to conducting a complete study, this model-migration method could significantly reduce the experimental efforts and facilitate the development process.
Conclusion: In conclusion, ESCAR demonstrated its applicability in emulating the in vivo SC environment and understanding the migration of protein-based therapeutics in the SC extracellular matrix. The future work would be focused on further optimizing ESCAR and expanding its applications via assessing more types of molecules and formulations. ESCAR had important implications for the research & development of SC large molecule (e.g., protein) formulations.
.jpg)
Molecular dynamics simulation and Computational Fluid Dynamics simulation of the developed system. (a) MD simulation between BSA and HA monomers (left); 8 identified residues labeled in red (right); (b) CFD simulation of the fluid flow inside the "SC" chamber
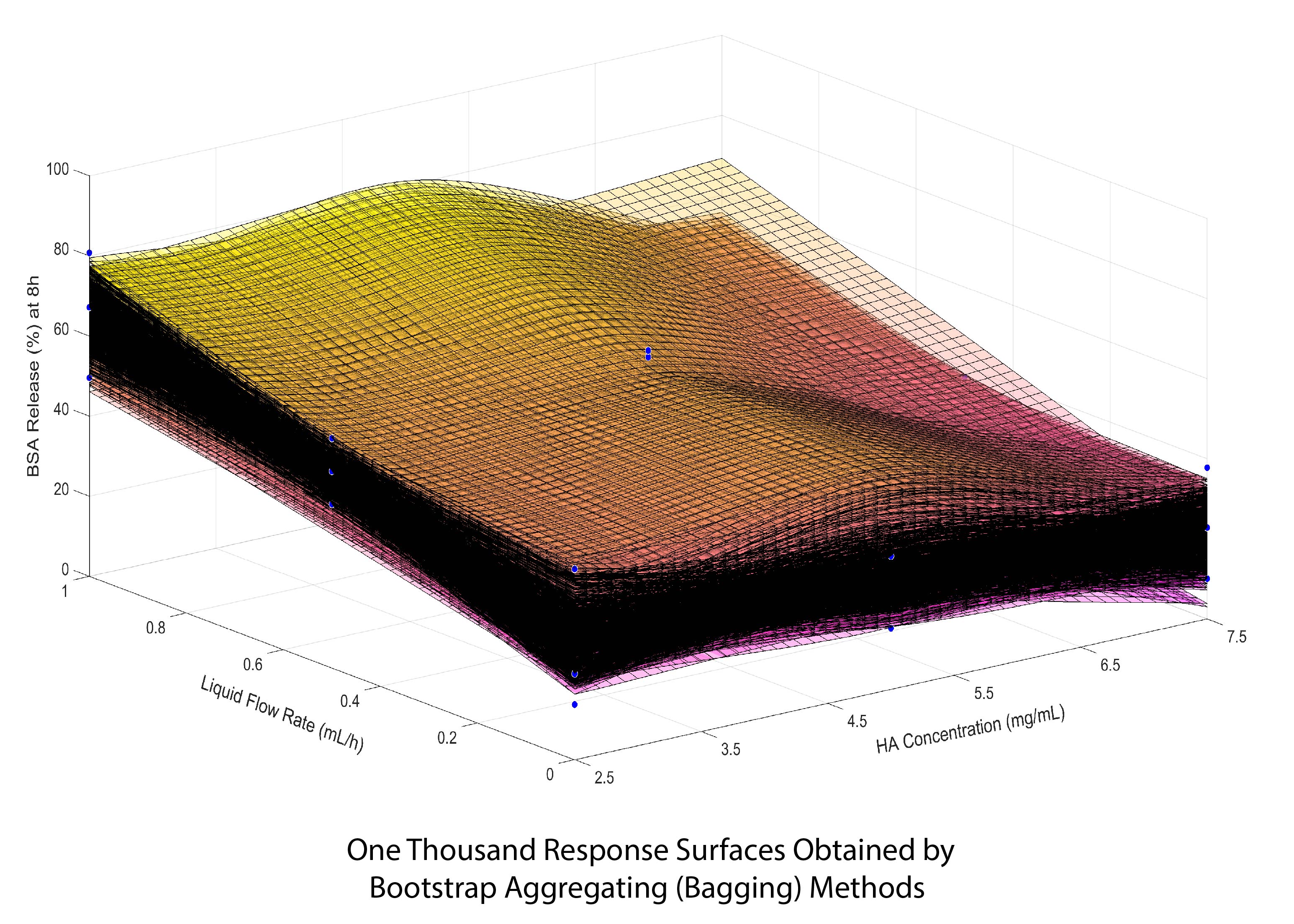
The modeling of drug release using response surfaces based on the multivariate spline interpolation and Bagging method
.jpg)
Model migration strategy for the development of a new model during process/formulation change. (a) release profiles of 30-mg dose and 15-mg dose; (b) schematic model migration methodology; (c) comparison between the base model and the complete model; (d) comparison between the migrated model and the complete model
Formulation and Delivery - Biomolecular - Drug Delivery, Devices, and Drug Device
Category: Poster Abstract
(M0930-07-39) Developing an In Vitro System to Emulate the In Vivo Subcutaneous Environment and Predict the Fate of Drugs in Subcutaneous Delivery: Assessment of Biologics
Monday, October 17, 2022
9:30 AM – 10:30 AM ET
- HL
Hao Lou, Ph.D.
University of Kansas
Lawrence, Kansas, United States - HL
Hao Lou, Ph.D.
University of Kansas
Lawrence, Kansas, United States
Presenting Author(s)
Main Author(s)
Purpose: We recently developed a prototype of an in vitro system, namely Emulator of SubCutaneous Absorption and Release (ESCAR), to emulate the in vivo subcutaneous (SC) environment. This study aimed to evaluate the ESCAR’s applicability in the assessment of biologics using a model protein bovine serum albumin (BSA).
Methods: ESCAR was fabricated by 3D printing technique with the aid of Computer-Aided Design (CAD). A DoE study was conducted to investigate the effects of fluid flow and hyaluronic acid (HA) concentration on drug (BSA) release. Molecular dynamics (MD) simulation was applied to study the electrostatic interaction between HA and BSA. The computational fluid dynamics (CFD) method was utilized to simulate the fluid flow inside ESCAR. Further, to reduce the experimental efforts during process/formulation change, a model migration method based on Bayesian inference was developed.
Results: The BSA release in HA solution was significantly slower than that in PBS, which was likely attributed to the following reasons: (1) lower viscosity in the medium; (2) higher resistance of BSA to diffusing through the characteristic length-scale of polymer chains; (3) electrostatic interaction between BSA and HA. Throughout the MD simulation, a total of 8 positively-charged residues (either arginine or lysine) in the BSA sequence were identified to have strong interactions with HA. Therefore, to reduce the interaction, scientists may consider mutating these residues to some neutral amino acids. Further, the fluid velocity and direction could be simulated by the CFD method. The results suggested that fluid flow/convection could substantially enhance the BSA release. To interpret this result from the in vivo perspective, the higher fluid flow/convection could lead to a faster migration from the SC injection site to the drainage site (e.g., lymphatic capillaries for most large molecules such as proteins). The DoE study pointed out that both fluid flow and HA concentration are critical parameters for the BSA release. The response surfaces based on the multivariate spline interpolation and Bagging method were developed to model the relationship between the input factors and the outputs (e.g., BSA release % at 2-hr, 4-hr, 8-hr, etc.). Further, a Bayesian interference-based method successfully migrated the model that was built in the old dataset (e.g., the dataset of 15-mg dose) to become a new model for the new dataset (e.g., the dataset of 30-mg dose). Compared to conducting a complete study, this model-migration method could significantly reduce the experimental efforts and facilitate the development process.
Conclusion: In conclusion, ESCAR demonstrated its applicability in emulating the in vivo SC environment and understanding the migration of protein-based therapeutics in the SC extracellular matrix. The future work would be focused on further optimizing ESCAR and expanding its applications via assessing more types of molecules and formulations. ESCAR had important implications for the research & development of SC large molecule (e.g., protein) formulations.
.jpg)
Molecular dynamics simulation and Computational Fluid Dynamics simulation of the developed system. (a) MD simulation between BSA and HA monomers (left); 8 identified residues labeled in red (right); (b) CFD simulation of the fluid flow inside the "SC" chamber

The modeling of drug release using response surfaces based on the multivariate spline interpolation and Bagging method
.jpg)
Model migration strategy for the development of a new model during process/formulation change. (a) release profiles of 30-mg dose and 15-mg dose; (b) schematic model migration methodology; (c) comparison between the base model and the complete model; (d) comparison between the migrated model and the complete model
