Back
Tumor Biology, Biomarkers, and Pathology
47: Epigenetic, transcriptional, and compositional shifts in clear cell renal cell carcinomas
Location: Poster Hall, Board E7
Width: 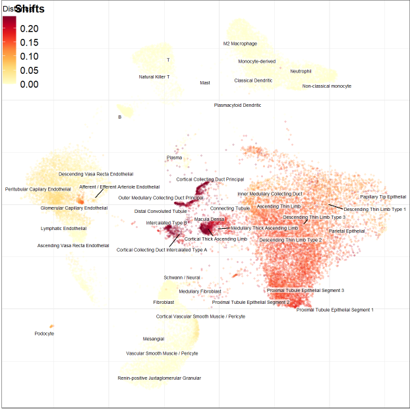
Compositional shifts in cell proportions between tumor and normal-adjacent samples


Lucas A. Salas, MD PhD MPH
Assistant Professor
Geisel School of Medicine at Dartmouth
Lebanon, NH, United States
Poster Presenter(s)
Background: Clear cell renal cell carcinoma (ccRCC) is a group of tumors characterized by alterations in chromosome 3p, 90% of them showing alterations in the VHL gene. Here we aim to evaluate biobanked samples to characterize single-cell epigenetic, transcriptional, and compositional shifts in tumor vs normal-adjacent samples.
Methods: We analyzed 57 tumor samples and six normal-adjacent kidney samples from patients in the Dartmouth Renal Tumors Biobank collected between 1994 and 2009. Samples were mechanically and enzymatically disassociated and preserved at -80 Celsius until processing. Cells were processed using the 10X multiome protocol. RNA counts and Chromatin accessibility peaks were extracted using Seurat.
Results: The mean age was 61.7 yrs (SD: 12.5), with 67% being stages I and II. 85,771 cell samples were analyzed. Relative compositional shifts were observed with an increased proportion in the tumors vs. normal adjacent in immune cells (22 vs. 15%), descending and ascending thin limbs (36 vs. 23%, and 2 vs. ~0%), fibroblasts (22 vs. 15%) pericytes (13 vs. 5%) and T cells (7 vs. 1.8%).
37% of all the cells captured in the tumor were aneuploid, predominantly from descending thin limb (24% of all cells). We observed a shift in the tumor's proportions of ascending thin limb cells and an increase in the cortical thick ascending limb cells in the normal-adjacent samples. After adjusting for sample cell compositions, we calculated gene programs. The top program showed 125 genes related to chromatin binding and organization, 2-oxoglutarate, and dioxygenase activity.
Conclusions: Distributional shifts with alterations in cell composition, gene expression, and chromatin accessibility of the ccRCC samples were observed. Predominant tumor cells were the descending thin limb, and aneuploid descending thin limb cells were captured in both the tumor and normal-adjacent samples. Altered programs were observed related to aneuploid cells.
Methods: We analyzed 57 tumor samples and six normal-adjacent kidney samples from patients in the Dartmouth Renal Tumors Biobank collected between 1994 and 2009. Samples were mechanically and enzymatically disassociated and preserved at -80 Celsius until processing. Cells were processed using the 10X multiome protocol. RNA counts and Chromatin accessibility peaks were extracted using Seurat.
Results: The mean age was 61.7 yrs (SD: 12.5), with 67% being stages I and II. 85,771 cell samples were analyzed. Relative compositional shifts were observed with an increased proportion in the tumors vs. normal adjacent in immune cells (22 vs. 15%), descending and ascending thin limbs (36 vs. 23%, and 2 vs. ~0%), fibroblasts (22 vs. 15%) pericytes (13 vs. 5%) and T cells (7 vs. 1.8%).
37% of all the cells captured in the tumor were aneuploid, predominantly from descending thin limb (24% of all cells). We observed a shift in the tumor's proportions of ascending thin limb cells and an increase in the cortical thick ascending limb cells in the normal-adjacent samples. After adjusting for sample cell compositions, we calculated gene programs. The top program showed 125 genes related to chromatin binding and organization, 2-oxoglutarate, and dioxygenase activity.
Conclusions: Distributional shifts with alterations in cell composition, gene expression, and chromatin accessibility of the ccRCC samples were observed. Predominant tumor cells were the descending thin limb, and aneuploid descending thin limb cells were captured in both the tumor and normal-adjacent samples. Altered programs were observed related to aneuploid cells.
