Back
Basic Science
9: DNA methylation-based tumor microenvironment deconvolution in clear cell renal cell carcinoma reveals significant survival association with specific cell types
Location: Poster Hall, Board A1
Width: 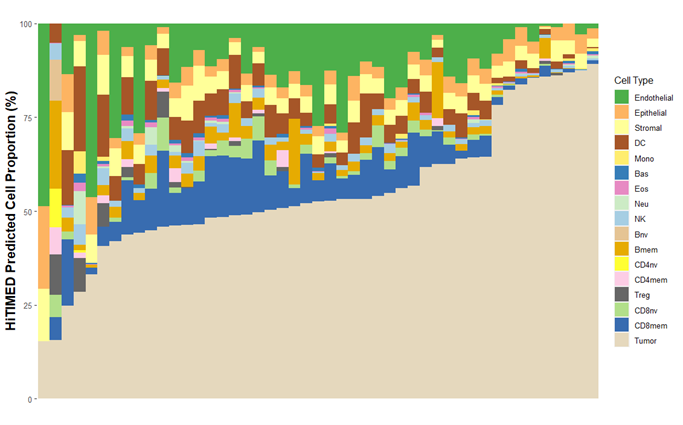
Figure 1. ccRCC tumor microenvironment cell composition deconvolved by HiTIMED


Ze Zhang
PhD Candidate
Dartmouth College
Hanover, NH, United States
Poster Presenter(s)
Background: Our group recently developed a DNA methylation-based bioinformatic tool, HiTIMED, to deconvolve the tumor microenvironment (TME) with high resolution, accuracy, and specificity. Seventeen cell types from three major TME components (tumor, immune, angiogenic) can be estimated. Here, we applied HiTIMED to clear cell renal cell carcinoma (ccRCC) samples to investigate the association between TME cell composition and survival outcome.
Methods: 47 ccRCC samples from the Dartmouth Renal Tumor Biobank were selected for a pilot study. Two hundred samples are aimed for the final study. Tandem bisulfite and oxidative-bisulfite conversion on DNA followed by hybridization to Infinium MethylationEPIC BeadChip were used to measure methylation and hydroxymethylation. HiTIMED was used to deconvolve the cell composition in ccRCC TME (Figure 1). Kaplan-Meier survival curves and Cox proportional hazard models adjusting for age, sex, and tumor stage were employed for survival analyses. Methylation dysregulation index (MDI) and hydroxymethylation dysregulation index (hMDI) were calculated.
Results: A higher than the median level of stromal cell proportion (>4.38% vs. ≤ 4.38%) is significantly associated with a better survival rate (HR: 0.49, 95% CI: [0.25, 0.96]). A higher than median level of CD4 memory cell proportion (>0% vs. =0%) is significantly associated with a worse survival outcome (HR: 2.16, 95% CI: [1.14, 4.09]). A higher than the median level of T regulatory cell proportion (>0% vs. =0%) is associated with a worse survival rate (HR: 2.09, 95% CI: [1.11, 3.94]). MDI and hMDI demonstrated increasing trends with the ccRCC tumor stage.
Conclusions: DNA methylation-based estimation of cell proportions in ccRCC TME revealed stromal, CD4 memory, and T regulatory cells significantly associated with survival outcomes. The observation promises future investigations on biological implications and roles of those cells in ccRCC carcinogenesis and progression.
Methods: 47 ccRCC samples from the Dartmouth Renal Tumor Biobank were selected for a pilot study. Two hundred samples are aimed for the final study. Tandem bisulfite and oxidative-bisulfite conversion on DNA followed by hybridization to Infinium MethylationEPIC BeadChip were used to measure methylation and hydroxymethylation. HiTIMED was used to deconvolve the cell composition in ccRCC TME (Figure 1). Kaplan-Meier survival curves and Cox proportional hazard models adjusting for age, sex, and tumor stage were employed for survival analyses. Methylation dysregulation index (MDI) and hydroxymethylation dysregulation index (hMDI) were calculated.
Results: A higher than the median level of stromal cell proportion (>4.38% vs. ≤ 4.38%) is significantly associated with a better survival rate (HR: 0.49, 95% CI: [0.25, 0.96]). A higher than median level of CD4 memory cell proportion (>0% vs. =0%) is significantly associated with a worse survival outcome (HR: 2.16, 95% CI: [1.14, 4.09]). A higher than the median level of T regulatory cell proportion (>0% vs. =0%) is associated with a worse survival rate (HR: 2.09, 95% CI: [1.11, 3.94]). MDI and hMDI demonstrated increasing trends with the ccRCC tumor stage.
Conclusions: DNA methylation-based estimation of cell proportions in ccRCC TME revealed stromal, CD4 memory, and T regulatory cells significantly associated with survival outcomes. The observation promises future investigations on biological implications and roles of those cells in ccRCC carcinogenesis and progression.
