Back
Poster, Podium & Video Sessions
Podium
PD21: Infections/Inflammation/Cystic Disease of the Genitourinary Tract: Kidney & Bladder II
PD21-12: Microbiota and Metabolites are Consistently Present on Implanted Urological Devices and Differ Significantly Based on Infection Status and Device Type
Saturday, May 14, 2022
8:50 AM – 9:00 AM
Location: Room 243
Glenn T. Werneburg*, Ava Adler, Sandip Vasavada, Howard Goldman, Raymond Rackley, Daniel Hettel, Bradley Gill, Kenneth Angermeier, Hadley Wood, Petar Bajic, Smita De, Daniel Shoskes, Jacqueline Zillioux, Aaron Miller, Cleveland, OH

Glenn T. Werneburg, MD, PHD
Cleveland Clinic
Podium Presenter(s)
Introduction: Urology employs multiple implanted devices, including sacral neuromodulators (SNM), artificial urinary sphincters (AUS), ureteral stents, and inflatable penile prostheses (IPP), to effectively modulate genitourinary function. A subset of urologic devices is colonized by microbes in the form of biofilms, but it is unknown why some, but not all, of these devices lead to clinical infection. We sought to determine the device-associated microbiota and metabolites, and assess and reconstitute biofilm formation of device isolates in vitro, to gain better understanding of underlying pathophysiology. We hypothesized that each device type would have a specific microbe and metabolite profile.
Methods: Patients scheduled to undergo removal/revision of devices (SNM, AUS, ureteral stent, IPP) for any cause were consented per IRB-approved protocol. First accessed portions of the devices were swabbed intraoperatively, with safeguards to avoid contamination. The swab samples, alongside controls, were subjected to next-generation sequencing and metabolomics to determine their composition and diversity. Microbes from devices were isolated and identified, and their biofilm-formation capacity was assessed using a semi-quantitative assay.
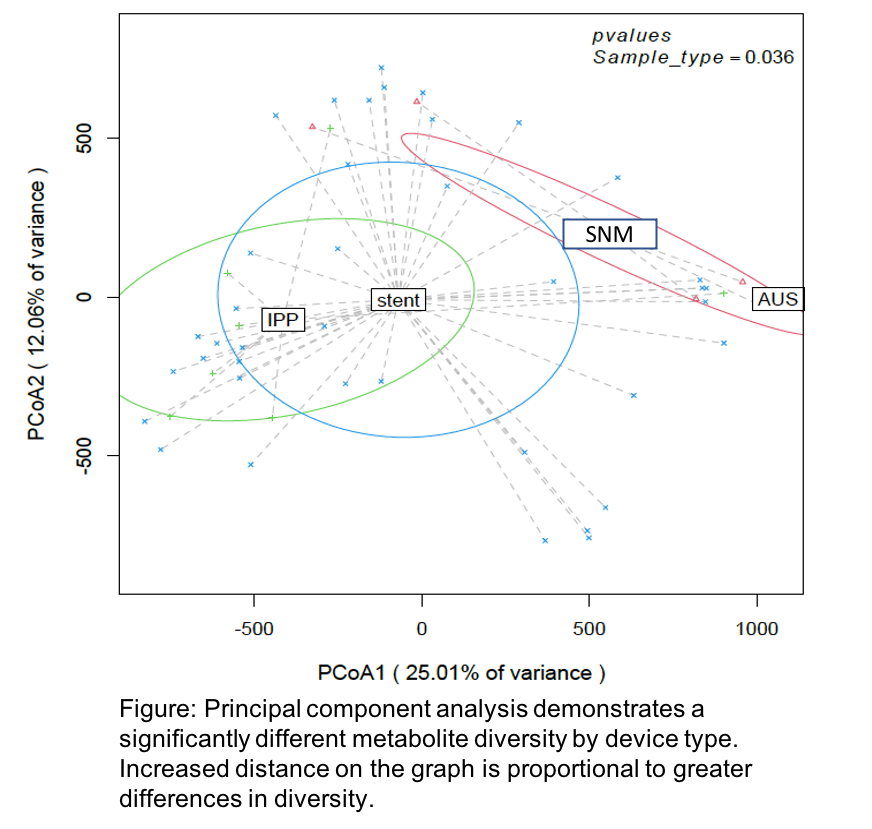
Results: 112 device swabs were analyzed by next-generation sequencing and metabolomics, and all samples harbored a detectable microbiome. The most common phylum was Proteobacteria. Diversity of microbiota (p=0.045) and metabolites (p=0.036) (Figure) differed significantly by device type. Microbial diversity differed between infected and noninfected devices (p=0.016). Of 32 isolates cultured, identified, and screened for biofilm formation, 13 exhibited biofilm formation in vitro including Escherichia coli, Enterococcus faecalis, and Staphylococcus epidermidis.
Conclusions: Devices consistently harbored bacteria. Bacterial and metabolite composition differed significantly by device type and infection status. Biofilm formation of isolates including pathogens was reconstitutable in vitro. Our results inform future studies, which will investigate device microbial composition and contributions to clinical infection and determine how bacteria isolated from devices impact host physiology.
Source of Funding: N/A
Methods: Patients scheduled to undergo removal/revision of devices (SNM, AUS, ureteral stent, IPP) for any cause were consented per IRB-approved protocol. First accessed portions of the devices were swabbed intraoperatively, with safeguards to avoid contamination. The swab samples, alongside controls, were subjected to next-generation sequencing and metabolomics to determine their composition and diversity. Microbes from devices were isolated and identified, and their biofilm-formation capacity was assessed using a semi-quantitative assay.
Results: 112 device swabs were analyzed by next-generation sequencing and metabolomics, and all samples harbored a detectable microbiome. The most common phylum was Proteobacteria. Diversity of microbiota (p=0.045) and metabolites (p=0.036) (Figure) differed significantly by device type. Microbial diversity differed between infected and noninfected devices (p=0.016). Of 32 isolates cultured, identified, and screened for biofilm formation, 13 exhibited biofilm formation in vitro including Escherichia coli, Enterococcus faecalis, and Staphylococcus epidermidis.
Conclusions: Devices consistently harbored bacteria. Bacterial and metabolite composition differed significantly by device type and infection status. Biofilm formation of isolates including pathogens was reconstitutable in vitro. Our results inform future studies, which will investigate device microbial composition and contributions to clinical infection and determine how bacteria isolated from devices impact host physiology.
Source of Funding: N/A


.jpg)
.jpg)