Back
Poster, Podium & Video Sessions
Moderated Poster
MP05: Stone Disease: Basic Research & Pathophysiology
MP05-06: Genetic Analysis of Kidney Stone Disease in a Multi-ethnic Cohort: Insights from Genome-Wide and Phenome-Wide Association Studies
Friday, May 13, 2022
8:45 AM – 10:00 AM
Location: Room 225
Parth Patel*, Vidhya Venkateswaran, Bogdan Pasanuic, Kymora Scotland, Los Angeles, CA
- PP
Parth M. Patel, MD
Urology Resident
UCLA
Poster Presenter(s)
Introduction: Nephrolithiasis, a common global ailment with significant clinical and economic health burden, has demonstrated heritability up to 56%. Various single nucleotide polymorphisms (SNPs) have been implicated in the development of kidney stone disease. Existing literature regarding this association is primarily in the form of genome-wide association studies or based on geographically limited samples. This genome- and phenome-wide association study aimed to evaluate the link between SNPs and kidney stone disease in a database of patients representing diverse ancestral backgrounds.
Methods: We retrospectively studied our repository of more than 27000 genotyped patients and their de-identified electronic health records. ICD-10 codes were aggregated to provide clinically meaningful groupings called phecodes. Genome-wide association testing was performed between all genotyped SNPs and phecodes associated with urolithiasis. We also tested the association between selected SNPs from our genome-wide association analysis and 1330 phecodes in the biobank to ascertain other phenotypic associations.
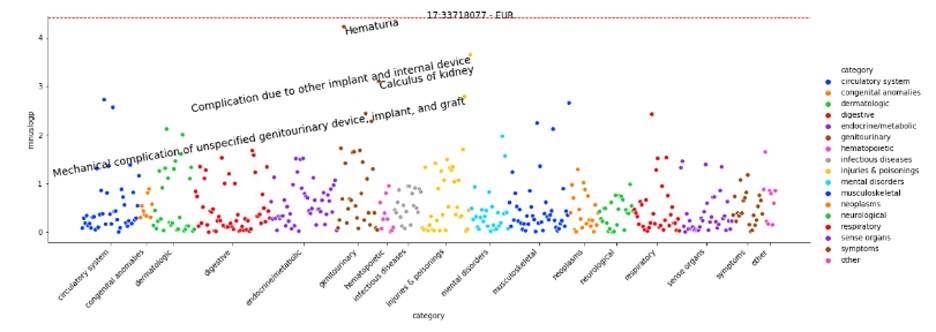
Results: The ancestry breakdown of these patients was 72.8% European, 16.3% Admixed American, 7.41% Eastern Asian, and 3.45% African. The lead SNPs from the trans-ancestry meta-analysis were rs2263090 and rs79616469 with p-values approaching genome-wide significance threshold of 5e-08 (Figure 1). In the phenome-wide association analysis, SNP rs2263090 showed associations with other kidney-related traits such as hematuria in EUR (Figure 2).
Conclusions: While numerous polymorphisms have been associated with kidney stone disease, our genome-wide and phenome-wide association studies suggest that their phenotypic expression may not be equal across genetic ancestral groupings. This promotes the concept that inheritable causes of nephrolithiasis are likely polygenic and their expression multifactorial, and much still remains to be learned about the molecular basis of kidney stone formation.
Source of Funding: N/A

Methods: We retrospectively studied our repository of more than 27000 genotyped patients and their de-identified electronic health records. ICD-10 codes were aggregated to provide clinically meaningful groupings called phecodes. Genome-wide association testing was performed between all genotyped SNPs and phecodes associated with urolithiasis. We also tested the association between selected SNPs from our genome-wide association analysis and 1330 phecodes in the biobank to ascertain other phenotypic associations.
Results: The ancestry breakdown of these patients was 72.8% European, 16.3% Admixed American, 7.41% Eastern Asian, and 3.45% African. The lead SNPs from the trans-ancestry meta-analysis were rs2263090 and rs79616469 with p-values approaching genome-wide significance threshold of 5e-08 (Figure 1). In the phenome-wide association analysis, SNP rs2263090 showed associations with other kidney-related traits such as hematuria in EUR (Figure 2).
Conclusions: While numerous polymorphisms have been associated with kidney stone disease, our genome-wide and phenome-wide association studies suggest that their phenotypic expression may not be equal across genetic ancestral groupings. This promotes the concept that inheritable causes of nephrolithiasis are likely polygenic and their expression multifactorial, and much still remains to be learned about the molecular basis of kidney stone formation.
Source of Funding: N/A



.jpg)
.jpg)