Back
Poster, Podium & Video Sessions
Moderated Poster
MP25: Trauma/Reconstruction/Diversion: Ureter (including Pyeloplasty) and Bladder Reconstruction (including fistula), Augmentation, Substitution, Diversion
MP25-16: Clinical Implications of the Urinary Diversion Microbiome after Radical Cystectomy
Saturday, May 14, 2022
10:30 AM – 11:45 AM
Location: Room 228
Saum Ghodoussipour, New Brunswick, CA, Seyedeh Sanam Ladi Seyedian*, David Nusbaum, Shane Pearce, Los Angeles, CA, Hiren Patel, Aishani Prem, Wendy Ullmer, Irvine, CA, Sumeet Bhanvadia, Hooman Djaladat, Anne Schuckman, Gangning Liang, Siamak Daneshmand, Los Angeles, CA

Seyedeh Sanam Ladi Seyedian, MD
University of Southern California
Poster Presenter(s)
Introduction: We use bacterial 16S rRNA and fungal ITS sequencing to identify the UD microbiome after radical and clinical implications of taxonomic and microbial diversity differences.
Methods: We enrolled patients who underwent RC and UD with either ileal conduit or neobladder under an IRB-approved protocol. A catheterized urine specimen from the native bladder and a swab of the ileal segment used to create the UD were collected in the operating room. We then collected sequential catheterized specimens from the UD until 1-year after surgery. Preoperative antibiotic prophylaxis was administered according to an institutional ERAS protocol. Microbial 16S rRNA and ITS sequencing and profiling were performed.
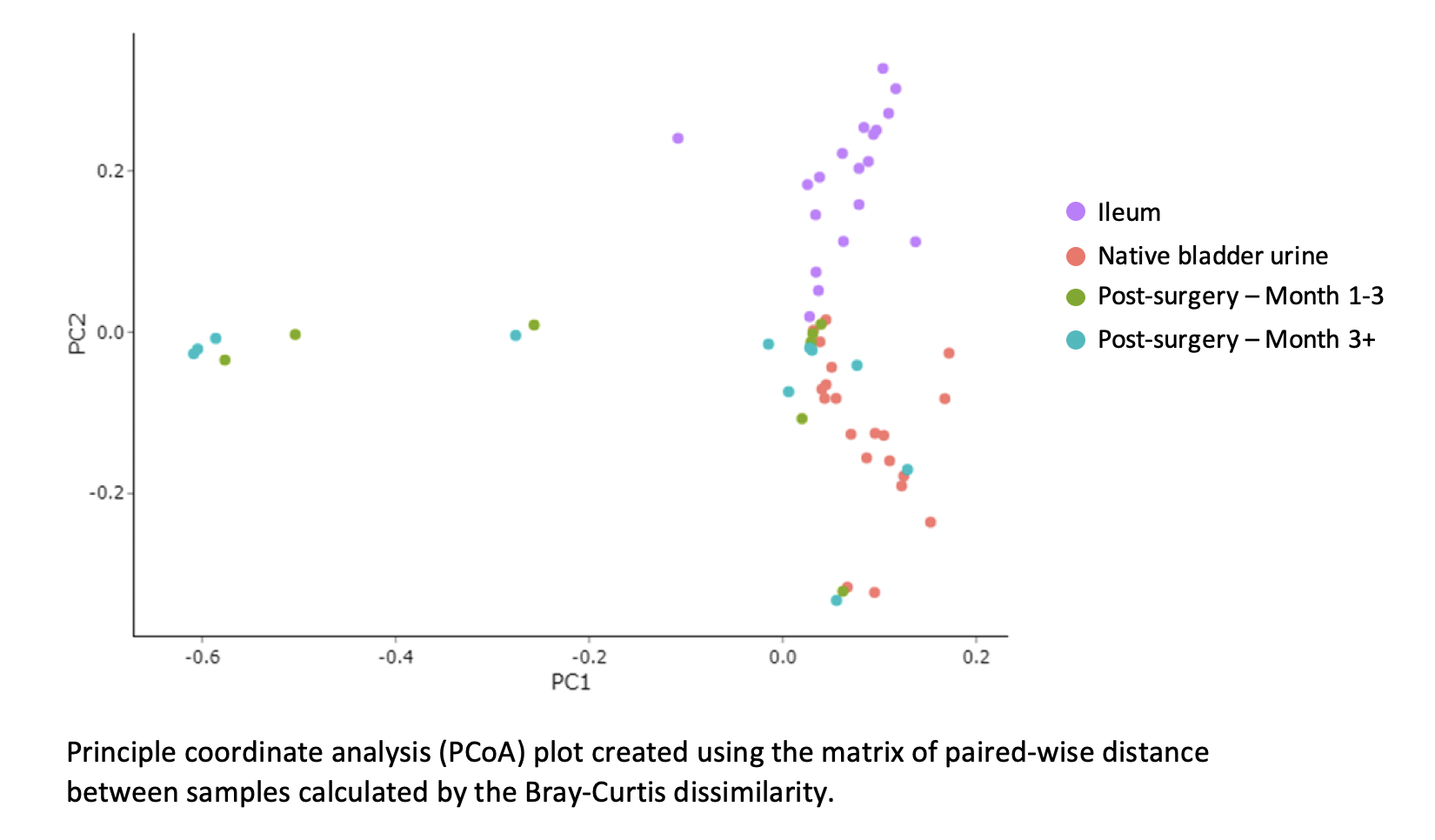
Results: We included 20 patients (75% male, 30% had neoadjuvant chemotherapy). The 90-day complication rate was 60% (n=12), including infectious in 25% (n=5). The average bacterial count, estimated by gene copy number, was higher in the bowel swab (1.38 x 108 cells/ml) than native bladder (7.23 x 106 cells/ml) as was alpha diversity, which measures the number of different species in each sample (mean species richness = 50 ± 26 in bowel and 29 ± 25 in native bladder). The average UD bacterial count and alpha diversity was high on post-operative day 1 (1.38 x 108 cells/ml and 31 ± 19, respectively), but over time, alpha diversity decreased while bacterial counts tended to increase (11 ± 12 and 4.54 x 108 cells/ml at 1 year, respectively). High fungal cell counts (>1,000 cells/ml) were noted in 7 patients and Candida species were abundant in all but one. Principal coordinate analysis showed microbial profiles of the native bladder and ileum clustering in distinct patterns, but the UD microbiome varied between patients (fig). Age, gender, receipt of neoadjuvant chemotherapy and type of diversion did not predict similarities in the UD microbiome. There were no significant differences in microbial diversity or bacterial count amongst patients with and without complications. However, a high fungal count was associated with any complication in the first month (p < 0.01) and first year (p < 0.05).
Conclusions: The UD microbiome differs from the native bladder and ileum and varies widely between patients. High fungal loads are associated with patients having any complication, potentially implicating Candida species as a pathogenic organism.
Source of Funding: Zymo Research Corp
Methods: We enrolled patients who underwent RC and UD with either ileal conduit or neobladder under an IRB-approved protocol. A catheterized urine specimen from the native bladder and a swab of the ileal segment used to create the UD were collected in the operating room. We then collected sequential catheterized specimens from the UD until 1-year after surgery. Preoperative antibiotic prophylaxis was administered according to an institutional ERAS protocol. Microbial 16S rRNA and ITS sequencing and profiling were performed.
Results: We included 20 patients (75% male, 30% had neoadjuvant chemotherapy). The 90-day complication rate was 60% (n=12), including infectious in 25% (n=5). The average bacterial count, estimated by gene copy number, was higher in the bowel swab (1.38 x 108 cells/ml) than native bladder (7.23 x 106 cells/ml) as was alpha diversity, which measures the number of different species in each sample (mean species richness = 50 ± 26 in bowel and 29 ± 25 in native bladder). The average UD bacterial count and alpha diversity was high on post-operative day 1 (1.38 x 108 cells/ml and 31 ± 19, respectively), but over time, alpha diversity decreased while bacterial counts tended to increase (11 ± 12 and 4.54 x 108 cells/ml at 1 year, respectively). High fungal cell counts (>1,000 cells/ml) were noted in 7 patients and Candida species were abundant in all but one. Principal coordinate analysis showed microbial profiles of the native bladder and ileum clustering in distinct patterns, but the UD microbiome varied between patients (fig). Age, gender, receipt of neoadjuvant chemotherapy and type of diversion did not predict similarities in the UD microbiome. There were no significant differences in microbial diversity or bacterial count amongst patients with and without complications. However, a high fungal count was associated with any complication in the first month (p < 0.01) and first year (p < 0.05).
Conclusions: The UD microbiome differs from the native bladder and ileum and varies widely between patients. High fungal loads are associated with patients having any complication, potentially implicating Candida species as a pathogenic organism.
Source of Funding: Zymo Research Corp


.jpg)
.jpg)