Back
Poster, Podium & Video Sessions
Moderated Poster
MP36: Renal Transplantation & Vascular Surgery
MP36-09: Validation of a urinary exosome mRNA signature for the diagnosis of human kidney transplant rejection
Sunday, May 15, 2022
7:00 AM – 8:15 AM
Location: Room 228
Rania El Fekih*, Boston, MA, Kurt Franzen, Brian Cole, James Hurley, Sonia Kumar, Johan Skog, Waltham, MA, Jamil Azzi, Boston, MA

Rania El Fekih, MD
Transplantation Research Center, Brigham and Women's Hospital
Poster Presenter(s)
Introduction: Early detection of kidney allograft rejection is crucial to avoid short and long-term adverse clinical outcomes. Gene transcripts detected in urinary exosomes can serve as a non-invasive predictor of acute kidney transplant rejection, and the stability of urinary exosomes makes them an ideal non-invasive diagnostic biomarker for kidney-transplant rejection.
Methods: Urine samples were collected from patients undergoing a transplant kidney biopsy either due to clinical indications or according to protocol. A total of 470 urine samples - split between training (329) and validation (141) cohorts - were collected. Exosomal RNA was isolated from 3-10ml urine using the ExosomeDx isolation platform ExoLution Transcript levels of 17 gene targets previously determined to be associated with kidney rejection (PMID: 33658284) were evaluated by RT-qPCR. Machine learning was applied to the training cohort data to determine an optimal algorithm for detecting kidney rejection.
Results: In this study, a total of 470 samples passed all quality control thresholds, and the rejection prevalence of 26.6%.
The cohort was divided into a 70% training partition, holding out 30% of samples for model evaluation. The partition was stratified by the rejection status, ensuring the same balance of rejection samples in both partitions.
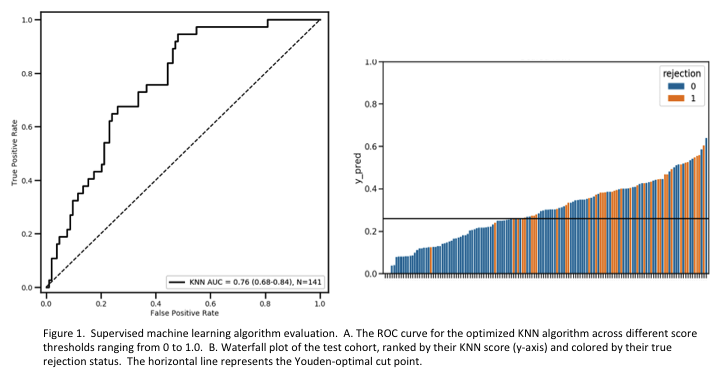
Each classifier was optimized by grid search before evaluation on the test set. The best performing model was a K-Nearest Neighbors (KNN) algorithm. This model attains an ROC AUC of 0.76 on the testing cohort. To predict the rejection status of samples, this algorithm finds the K nearest neighbors among the training cohort. The ratio of rejection samples to nonrejection samples among these nearest neighbors, weighted by their distance, is the prediction score, which ranges from 0 to 1.0. The optimal binary cutoff was determined using the Youden index (cutoff = 0.259). Samples with a KNN score above this cutoff are determined to have ongoing rejection. The negative predictive value was 0.96, providing high confidence that a sample with a negative prediction is truly negative.
Conclusions: RNA signatures derived from urinary exosomes represent a powerful and non-invasive tool to assess kidney allograft rejection and support clinicians in therapeutic decisions.
Source of Funding: Exosomedx, Biotechne
Methods: Urine samples were collected from patients undergoing a transplant kidney biopsy either due to clinical indications or according to protocol. A total of 470 urine samples - split between training (329) and validation (141) cohorts - were collected. Exosomal RNA was isolated from 3-10ml urine using the ExosomeDx isolation platform ExoLution Transcript levels of 17 gene targets previously determined to be associated with kidney rejection (PMID: 33658284) were evaluated by RT-qPCR. Machine learning was applied to the training cohort data to determine an optimal algorithm for detecting kidney rejection.
Results: In this study, a total of 470 samples passed all quality control thresholds, and the rejection prevalence of 26.6%.
The cohort was divided into a 70% training partition, holding out 30% of samples for model evaluation. The partition was stratified by the rejection status, ensuring the same balance of rejection samples in both partitions.
Each classifier was optimized by grid search before evaluation on the test set. The best performing model was a K-Nearest Neighbors (KNN) algorithm. This model attains an ROC AUC of 0.76 on the testing cohort. To predict the rejection status of samples, this algorithm finds the K nearest neighbors among the training cohort. The ratio of rejection samples to nonrejection samples among these nearest neighbors, weighted by their distance, is the prediction score, which ranges from 0 to 1.0. The optimal binary cutoff was determined using the Youden index (cutoff = 0.259). Samples with a KNN score above this cutoff are determined to have ongoing rejection. The negative predictive value was 0.96, providing high confidence that a sample with a negative prediction is truly negative.
Conclusions: RNA signatures derived from urinary exosomes represent a powerful and non-invasive tool to assess kidney allograft rejection and support clinicians in therapeutic decisions.
Source of Funding: Exosomedx, Biotechne


.jpg)
.jpg)