Back
Poster, Podium & Video Sessions
Moderated Poster
MP37: Infections/Inflammation/Cystic Disease of the Genitourinary Tract: Prostate & Genitalia
MP37-08: Urinary Microbiome as a Biomarker for Prostate Cancer
Sunday, May 15, 2022
7:00 AM – 8:15 AM
Location: Room 225
Michael Liss*, John Lee, San Antonio, TX, James White, Baltimore, MD, Teresa Johnson-Pais, Zhao Lai, San Antonio, TX, Dean Troyer, Norfolk, VA, Robin Leach, Brian Wickes, San Antonio, TX

Michael A. Liss, MD,PhD,FACS,MS
University of Texas Health San Antonio
Poster Presenter(s)
Introduction: To develop a diagnostic biomarker development strategy to investigate potential microbial biomarkers for prostate cancer.
Methods: We designed a prospective training cohort with a case-control design to create a microbiome-based biomarker. In the case group, we enrolled men that went to surgery. We collected urine before prostate biopsy, prostatectomy tissue, and urine after prostatectomy in the same patients to perform paired filtering to identify the most important microbiota (42 samples, n=14). In the control group, we collected urine from normal men with very low risk of prostate cancer (PSA <1.0, screened for more than 10 years) before and after prostate exam (44 samples, n=22). We used a shotgun metagenomic sequencing at 6M and single end 150 bp using Illumina HiSeq platform. We tested the biomarker in a prospectively enrolled validation cohort undergoing prostate biopsy (n=45). Our primary outcome clinically significant prostate cancer (ISUP Group 2). We additionally validated the 24 microbial taxa in TCGA prostate tissue and a proteomics cohort.
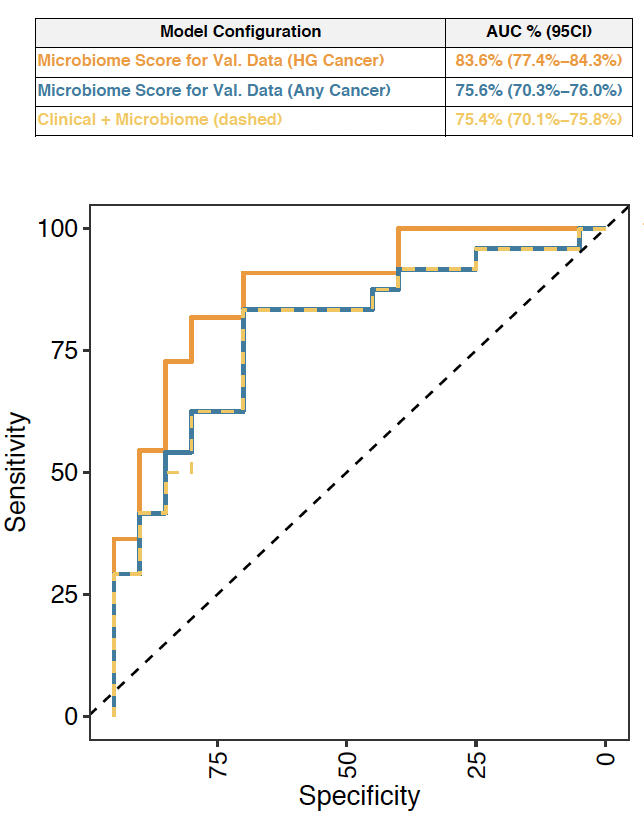
Results: Using our training cohort, we identified 24 candidate microbial taxa after performing paired comparisons and contaminant removal strategies. Using all microbes, the median area under the receiver operator curve (AUC) using 100 iterations of random subsampling was 0.87 (95% CI 0.81-0.92). We subsequently used a prostate biopsy validation cohort to test the best performing model. We identified eight microbial taxa that had an AUC of 0.84 (95% CI 0.74-0.85) that outperformed clinical risk factors including PSA (Figure 1). In the figure, we show the performance of the biomarker in the validation cohort (VAL) performance in high grade cancer (HG=ISUP>1), any cancer, and showed no improvement in AUC performance when clinical factors were added. We cross-referenced our 24 taxa to a published 187 microbiome taxa obtained from The Cancer Genome Atlas (TCGA) noting 19 (19/24, 76%) matches. We also investigated the 24 taxa within our existing proteomic data derived from mass spectrometry based proteomic analysis a previous cohort of 87 men on active surveillance confirming 8 of the 24 taxa.
Conclusions: Using a unique cohort to filter bacterial signatures for prostate cancer, we identified eight bacterial taxa associated with cancer that performed well in a validation cohort.
Source of Funding: Los Padres Foundation
Methods: We designed a prospective training cohort with a case-control design to create a microbiome-based biomarker. In the case group, we enrolled men that went to surgery. We collected urine before prostate biopsy, prostatectomy tissue, and urine after prostatectomy in the same patients to perform paired filtering to identify the most important microbiota (42 samples, n=14). In the control group, we collected urine from normal men with very low risk of prostate cancer (PSA <1.0, screened for more than 10 years) before and after prostate exam (44 samples, n=22). We used a shotgun metagenomic sequencing at 6M and single end 150 bp using Illumina HiSeq platform. We tested the biomarker in a prospectively enrolled validation cohort undergoing prostate biopsy (n=45). Our primary outcome clinically significant prostate cancer (ISUP Group 2). We additionally validated the 24 microbial taxa in TCGA prostate tissue and a proteomics cohort.
Results: Using our training cohort, we identified 24 candidate microbial taxa after performing paired comparisons and contaminant removal strategies. Using all microbes, the median area under the receiver operator curve (AUC) using 100 iterations of random subsampling was 0.87 (95% CI 0.81-0.92). We subsequently used a prostate biopsy validation cohort to test the best performing model. We identified eight microbial taxa that had an AUC of 0.84 (95% CI 0.74-0.85) that outperformed clinical risk factors including PSA (Figure 1). In the figure, we show the performance of the biomarker in the validation cohort (VAL) performance in high grade cancer (HG=ISUP>1), any cancer, and showed no improvement in AUC performance when clinical factors were added. We cross-referenced our 24 taxa to a published 187 microbiome taxa obtained from The Cancer Genome Atlas (TCGA) noting 19 (19/24, 76%) matches. We also investigated the 24 taxa within our existing proteomic data derived from mass spectrometry based proteomic analysis a previous cohort of 87 men on active surveillance confirming 8 of the 24 taxa.
Conclusions: Using a unique cohort to filter bacterial signatures for prostate cancer, we identified eight bacterial taxa associated with cancer that performed well in a validation cohort.
Source of Funding: Los Padres Foundation


.jpg)
.jpg)