Back
Poster, Podium & Video Sessions
Moderated Poster
MP43: Prostate Cancer: Localized: Active Surveillance
MP43-17: Predicting True Pathologic Gleason Grade During Active Surveillance for Prostate Cancer: Calibration of the Johns Hopkins ActiveCare Model in Five Large Active Surveillance Cohorts
Sunday, May 15, 2022
10:30 AM – 11:45 AM
Location: Room 222
Christian Pavlovich*, Baltimore, MD, Daan Nieboer, Rotterdam, Netherlands, Mufaddal Mamawala, Akihiko Nishimura, Patricia Landis, Katarzyna Macura, Jonathan Epstein, Baltimore, MD, Jozien Helleman, Rotterdam, Netherlands, Bruce Trock, Baltimore, MD, Movember Foundation’s Global Action Plan Prostate Cancer Active Surveillance (GAP3) consortium, Rotterdam, Netherlands

Christian Paul Pavlovich, MD
Professor of Urology
Johns Hopkins University School of Medicine
Poster Presenter(s)
Introduction: We developed a Bayesian latent state model that predicts true pathologic grade group (PGG) for men on active surveillance (AS) for low risk prostate cancer. We sought to recalibrate the current model to five external cohorts to assess accuracy and wider applicability.
Methods: Our “ActiveCare” model predicts PGG of individual AS patients from their demographic, longitudinal PSA, and serial biopsy data. This dynamic model is first trained on a dataset that includes patients whose PGG has been confirmed at radical prostatectomy (RP), then predicts unobserved, true PGG for other patients. We shared the model code with the Global Action Plan Prostate Cancer Active Surveillance (GAP3) initiative, which houses data from 27 AS cohorts (v3.3). Our model was first externally validated using data from five of the largest AS cohorts in GAP3 (Milan, MSKCC, MUSIC, PRIAS, UCSF), and then recalibrated to each. The ability of the recalibrated ActiveCare model to discriminate between patients with and without PGG >1 was assessed using the area under the ROC (AUC).
Results: The ActiveCare model was validated and calibrated to each of the five largest cohorts in GAP3 (in total 7,697 patients) (range: 768-3519). Median age of patients at diagnosis was 65 and median PSA ranged from 4.6-5.7. The percentage of AS patients who underwent RP ranged from 9%-26%. The percentage of patients with PGG > 1 at RP ranged from 64%-87%. At external validation, AUCs ranged from 0.5-0.6. However, by calibrating our model to each cohort, we achieved AUCs competitive with or exceeding those of other prognostic models to date (0.64-0.79) with the added benefit that our model predicts true PGG (rather than biopsy grade group).
Conclusions: Upon recalibration, our ActiveCare model was able to discriminate between PGG across disparate AS cohorts with reasonable ability. Further work to improve across-cohort calibration using hierarchical modeling to allow for borrowing of information across cohorts is underway.
Source of Funding: Movember and Osprey Foundations. The funders did not play any role in study design, collection, analysis or interpretation of data, nor in the drafting of this abstract.
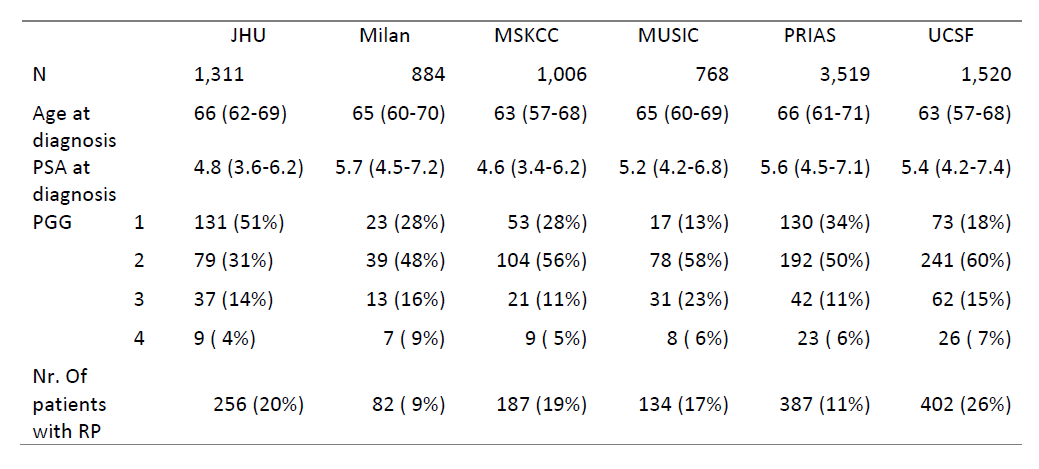

Methods: Our “ActiveCare” model predicts PGG of individual AS patients from their demographic, longitudinal PSA, and serial biopsy data. This dynamic model is first trained on a dataset that includes patients whose PGG has been confirmed at radical prostatectomy (RP), then predicts unobserved, true PGG for other patients. We shared the model code with the Global Action Plan Prostate Cancer Active Surveillance (GAP3) initiative, which houses data from 27 AS cohorts (v3.3). Our model was first externally validated using data from five of the largest AS cohorts in GAP3 (Milan, MSKCC, MUSIC, PRIAS, UCSF), and then recalibrated to each. The ability of the recalibrated ActiveCare model to discriminate between patients with and without PGG >1 was assessed using the area under the ROC (AUC).
Results: The ActiveCare model was validated and calibrated to each of the five largest cohorts in GAP3 (in total 7,697 patients) (range: 768-3519). Median age of patients at diagnosis was 65 and median PSA ranged from 4.6-5.7. The percentage of AS patients who underwent RP ranged from 9%-26%. The percentage of patients with PGG > 1 at RP ranged from 64%-87%. At external validation, AUCs ranged from 0.5-0.6. However, by calibrating our model to each cohort, we achieved AUCs competitive with or exceeding those of other prognostic models to date (0.64-0.79) with the added benefit that our model predicts true PGG (rather than biopsy grade group).
Conclusions: Upon recalibration, our ActiveCare model was able to discriminate between PGG across disparate AS cohorts with reasonable ability. Further work to improve across-cohort calibration using hierarchical modeling to allow for borrowing of information across cohorts is underway.
Source of Funding: Movember and Osprey Foundations. The funders did not play any role in study design, collection, analysis or interpretation of data, nor in the drafting of this abstract.


.jpg)
.jpg)