Back
Poster, Podium & Video Sessions
Best Poster Award
MP45: Prostate Cancer: Markers
MP45-14: The spatial landscape of clonal somatic mutations in benign and malignant prostate epithelia
Sunday, May 15, 2022
1:00 PM – 2:15 PM
Location: Room 225
Andrew Erickson*, Oxford, United Kingdom, Emelie Berglund, Mengxiao He, Maja Marklund, Reza Mirzazadeh, Niklas Schultz, Ludvig Bergenstråhle, Linda Kvastad, Alma Andersson, Joseph Bergenstråhle, Ludvig Larsson, Solna, Sweden, Alia Shamikh, Elisa Basmaci, Teresita Diaz De Ståhl, Stockholm, Sweden, Timothy Rajakumar, Oxford, United Kingdom, Kim Thrane, Solna, Sweden, Andrew Ji, Paul Khavari, Stanford, CA, Firaz Tarish, Solna, Sweden, Anna Tanoglidi, Uppsala, Sweden, Jonas Maaskola, Solna, Sweden, Richard Colling, Oxford, United Kingdom, Tuomas Mirtti, Helsinki, Finland, Freddie C Hamdy, Dan Woodcock, Oxford, United Kingdom, Thomas Helleday, Solna, Sweden, Ian Mills, Oxford, United Arab Emirates, Alastair Lamb, Oxford, United Kingdom, Joakim Lundeberg, Solna, Sweden

Alastair Lamb, MD, FRCS
University of Oxford
Poster Presenter(s)
Introduction: Defining the transition from benign to malignant tissue is fundamental to improve early diagnosis of cancer. We aimed to conduct an unsupervised approach to study spatial genome integrity, in situ, to gain molecular insight into clonal relationships.
Methods: We employed spatially resolved transcriptomics (Visium, 10x Genomics) to infer spatial copy number variations in >120 000 spatial regions across multiple organs, tissues and tumors, including three whole axial prostates from patients with primary prostate cancer. We used this information to deduce clonal relationships between regions harboring 5-20 cells.
Results: We performed an in-depth spatial analysis of prostate cancer that includes an unprecedented interrogation of up to 50,000 tissue domains in a single patient, and 120,000 tissue domains across 10 patients. For these domains we inferred genome-wide information, which facilitated data driven clustering in a tissue wide fashion at high resolution.
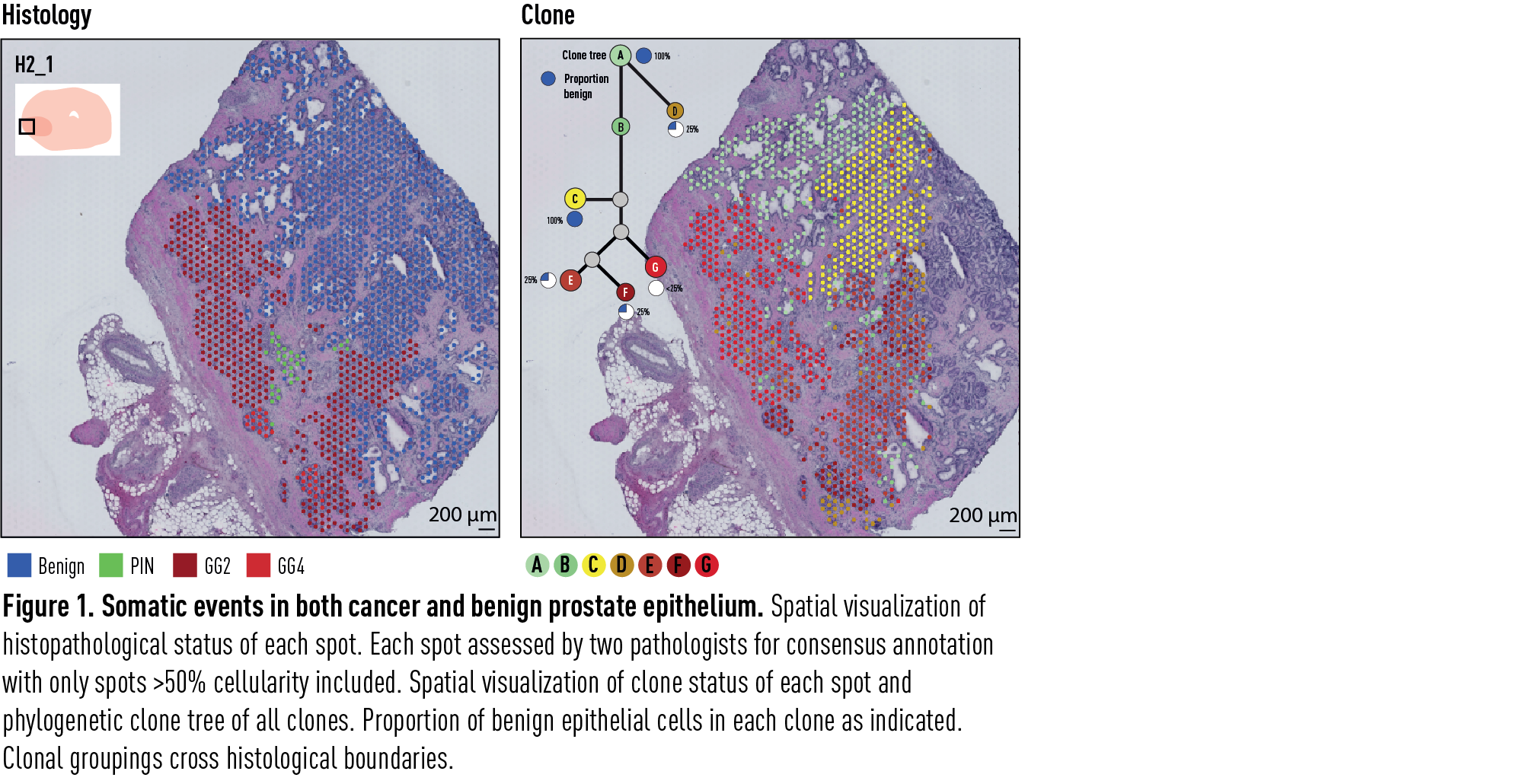
We demonstrated that genome-wide copy number variation reveals distinct clonal patterns within tumours and in nearby benign tissue. In section H2_1 (Figure 1), we observed that many CNVs occurred in clone C, most notably in chromosome 8. This clone constituted a region of exclusively benign acinar cells branching off a duct lined by largely copy neutral cells in clone A and B. Cancer clones E, F and G then arose from this clone with further mutational events.
Conclusions: In this analysis, we present the first large-scale, comprehensive atlas of genomic evolution at high spatial resolution in prostate cancer. Our results suggest a model for how genomic instability arises in histo-pathologically benign tissue that may represent early events in cancer evolution. Furthermore the spatial information allowed us to identify small clonal units not evident from morphology and which might therefore be overlooked by pathologists. We highlight the power of an unsupervised approach to capture the molecular and spatial continuums in a tissue context and challenge the rationale for focal therapy in prostate cancer.
Source of Funding: The Swedish Research Council, The Swedish Cancer Society, The Swedish Childhood Cancer Fund, Swedish Foundation for Strategic Research, Knut and Alice Wallenberg Foundation, Science for Life Laboratory/ Postdoc program, European Research Council Advanced Grant, Cancer Research UK Clinician Scientist Fellowship, John Black Charitable Foundation, Cancer Foundation Finland , Hospital District of Helsinki and Uusimaa, Academy of Finland, NIHR Oxford Biomedical Research Centre - Innovation & Evaluation Theme
Methods: We employed spatially resolved transcriptomics (Visium, 10x Genomics) to infer spatial copy number variations in >120 000 spatial regions across multiple organs, tissues and tumors, including three whole axial prostates from patients with primary prostate cancer. We used this information to deduce clonal relationships between regions harboring 5-20 cells.
Results: We performed an in-depth spatial analysis of prostate cancer that includes an unprecedented interrogation of up to 50,000 tissue domains in a single patient, and 120,000 tissue domains across 10 patients. For these domains we inferred genome-wide information, which facilitated data driven clustering in a tissue wide fashion at high resolution.
We demonstrated that genome-wide copy number variation reveals distinct clonal patterns within tumours and in nearby benign tissue. In section H2_1 (Figure 1), we observed that many CNVs occurred in clone C, most notably in chromosome 8. This clone constituted a region of exclusively benign acinar cells branching off a duct lined by largely copy neutral cells in clone A and B. Cancer clones E, F and G then arose from this clone with further mutational events.
Conclusions: In this analysis, we present the first large-scale, comprehensive atlas of genomic evolution at high spatial resolution in prostate cancer. Our results suggest a model for how genomic instability arises in histo-pathologically benign tissue that may represent early events in cancer evolution. Furthermore the spatial information allowed us to identify small clonal units not evident from morphology and which might therefore be overlooked by pathologists. We highlight the power of an unsupervised approach to capture the molecular and spatial continuums in a tissue context and challenge the rationale for focal therapy in prostate cancer.
Source of Funding: The Swedish Research Council, The Swedish Cancer Society, The Swedish Childhood Cancer Fund, Swedish Foundation for Strategic Research, Knut and Alice Wallenberg Foundation, Science for Life Laboratory/ Postdoc program, European Research Council Advanced Grant, Cancer Research UK Clinician Scientist Fellowship, John Black Charitable Foundation, Cancer Foundation Finland , Hospital District of Helsinki and Uusimaa, Academy of Finland, NIHR Oxford Biomedical Research Centre - Innovation & Evaluation Theme


.jpg)
.jpg)