Back
Poster, Podium & Video Sessions
Podium
PD12: Bladder Cancer: Basic Research & Pathophysiology II
PD12-04: Survey of B and Plasma Cells in Bladder Cancer Across Tumor Microenvironments
Friday, May 13, 2022
1:30 PM – 1:40 PM
Location: Room 244
Yuanshuo Wang*, New York, NY, Jorge Daza, Buffalo, NY, Emilie Grasset, New York, NY, Giuliana Magri, Barcelona, Spain, Daniel Ranti, Kristin Beaumont, Adam Farkas, Michelle Tran, Li Wang, Rachel Brody, Reza Mehrazin, Peter Wiklund, Robert Sebra, Jun Zhu, Matthew Galsky, Nina Bhardwaj, John Sfakianos, Amir Horowitz, New York, NY
- YW
Podium Presenter(s)
Introduction: Tumor infiltrating B cells in bladder cancer (BCa) have been associated with cancer invasion, but little is understood regarding their roles in different tumor microenvironments. In this study, we analyzed data from a large single cell (SC) RNA sequencing cohort of tumor (n=18) and PBMC (n=16) specimens from 26 BCa patients and PBMC samples from healthy donors (n=3) to profile B cells and plasma cells (PCs) in relation to BCa sample types.
Methods: SC data was imported into R (v4.03) using Seurat (v4.0.1). Genes expressed in fewer than 3 cells, cells with <200 or >2500 unique genes, and cells containing >15% mitochondrial genes were discarded. Data normalization, variable gene identification, integration, and scaling were performed. Known B cell and PC markers (MS4A1, IGHD, IGHM, IGHG1/2/3/4, IGHA1/2, MZB1, XBP1) were used to identify B cell and PC clusters after dimensionality reduction and graph-based clustering.
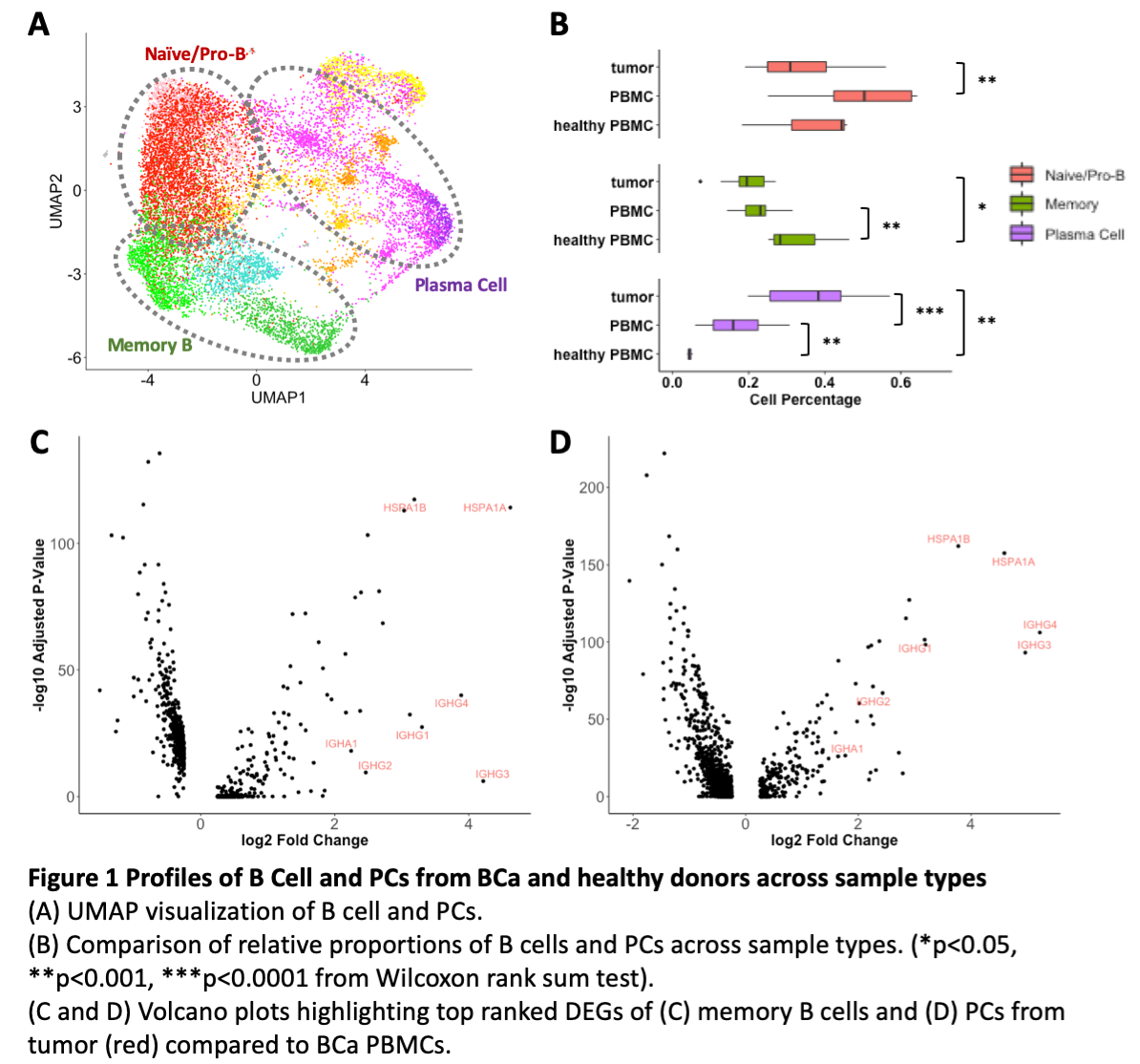
Results: We identified 15,008 naïve and memory B cells and PCs by their marker gene expression (Fig. 1A). Profiling these populations across sample types, we found a proportional decrease in memory B cells in BCa tumor and PBMC compared to healthy PBMCs (Fig. 1B). The high percentage of naïve B cells in patient PBMC suggest a possible stall in maturation to memory B cells, while the higher proportion of PCs in tumor suggest a possible competing maturation pathway between PCs and memory B cells from germinal center (GC) B cells. In addition, we observed differences in differentially expressed genes (DEGs) across sample types. Both memory B cells and PCs from BCa PBMC had significantly upregulated expression of ribosomal genes compared to healthy PBMC, and memory B cells and PCs also showed significantly higher expression of IGHG1/2/3/4 and IGHA1 in tumor relative to BCa PBMC (Fig. 1C-D).
Conclusions: In our study of a large BCa SC cohort, we observed significant proportional differences in B cell and PCs across BCa tumors, PBMCs, and healthy PBMCs that could possibly be attributed to a stall in maturation from naïve to memory B cell in the BCa blood or competing maturation pathways between PCs and memory B cells from GC B cells in BCa tumor. We also observed significant increases in genes supporting upregulation of immunoglobulin production both in tumor and in BCa blood relative to BCa PBMCs and healthy PBMCs, respectively.
Source of Funding: Institutional funds
Methods: SC data was imported into R (v4.03) using Seurat (v4.0.1). Genes expressed in fewer than 3 cells, cells with <200 or >2500 unique genes, and cells containing >15% mitochondrial genes were discarded. Data normalization, variable gene identification, integration, and scaling were performed. Known B cell and PC markers (MS4A1, IGHD, IGHM, IGHG1/2/3/4, IGHA1/2, MZB1, XBP1) were used to identify B cell and PC clusters after dimensionality reduction and graph-based clustering.
Results: We identified 15,008 naïve and memory B cells and PCs by their marker gene expression (Fig. 1A). Profiling these populations across sample types, we found a proportional decrease in memory B cells in BCa tumor and PBMC compared to healthy PBMCs (Fig. 1B). The high percentage of naïve B cells in patient PBMC suggest a possible stall in maturation to memory B cells, while the higher proportion of PCs in tumor suggest a possible competing maturation pathway between PCs and memory B cells from germinal center (GC) B cells. In addition, we observed differences in differentially expressed genes (DEGs) across sample types. Both memory B cells and PCs from BCa PBMC had significantly upregulated expression of ribosomal genes compared to healthy PBMC, and memory B cells and PCs also showed significantly higher expression of IGHG1/2/3/4 and IGHA1 in tumor relative to BCa PBMC (Fig. 1C-D).
Conclusions: In our study of a large BCa SC cohort, we observed significant proportional differences in B cell and PCs across BCa tumors, PBMCs, and healthy PBMCs that could possibly be attributed to a stall in maturation from naïve to memory B cell in the BCa blood or competing maturation pathways between PCs and memory B cells from GC B cells in BCa tumor. We also observed significant increases in genes supporting upregulation of immunoglobulin production both in tumor and in BCa blood relative to BCa PBMCs and healthy PBMCs, respectively.
Source of Funding: Institutional funds


.jpg)
.jpg)