Back
Poster, Podium & Video Sessions
Podium
PD36: Infertility: Basic Research & Pathophysiology
PD36-12: Abnormalities in Both Somatic and Germ Cells Differentiate NOA Testes from Normal Controls Using Single Cell RNA Sequencing
Sunday, May 15, 2022
8:50 AM – 9:00 AM
Location: Room 255
Arina Piechka*, Ghazal Ebrahimi, Meghan Robinson, Anton Afanassiev, Geoffrey Schiebinger, Eric Belanger, Faraz Hach, Ryan Flannigan, Vancouver, Canada
- AP
Podium Presenter(s)
Introduction: The etiology of Non-obstructive azoospemia (NOA) remains largely unknown. Furthermore, significant heterogeneity exists between and within NOA patients’ testes. This study aimed to capture heterogeneity within an NOA testis and investigate cell-type-specific abnormalities to normal controls.
Methods: Two testis biopsies were derived from one patient with NOA with known histologic heterogeneity, and one normal control. ScRNAseq was carried out using 10X Genomics and Illumina sequencing platforms in technical duplicates. Cell clustering, gene expression, Ingenuity Pathway Analysis (IPA), ligand-receptor analyses and TradeSeq pseudotime differential expression analyses were used to evaluate the datasets.
Results: Cell types detected in our normal control was congruent with the previously published dataset. One NOA sample largely lacked germ cells and was broadly categorized as Sertoli Cell Only (SCO). This sample uniquely demonstrated a large cluster of cells expressing a mixture of Leydig and myoid genes; this same cluster exhibited high expression of NR4A1–known marker of common progenitor of Leydig and myoid cells. T cells and granulocytes were also present while absent among normal controls. In the second NOA sample, we detected all germ cells up to elongated spermatids and labelled this as hypospermatogenesis (HS). In this HS NOA sample, T cells were also present. After aligning the normal germ cells and HS NOA germ cells in pseudo time, differential gene expression and IPA pathway analyses revealed that NOA-derived germ cells had down regulation in RNA and nucleotide base excision repair mechanisms suggesting intrinsic deficiencies. Histology corroborated our scRNAseq results.
Conclusions: Differentiating features of the SCO sample were the presence of T cells, granulocytes and an interstitial progenitor population. Pathway analyses of NOA germ cells suggest intrinsic abnormalities in RNA processing.
Source of Funding: Canadian Institutes of Health Research; American Society of Reproductive Medicine; Vancouver Coastal Health Research Institute
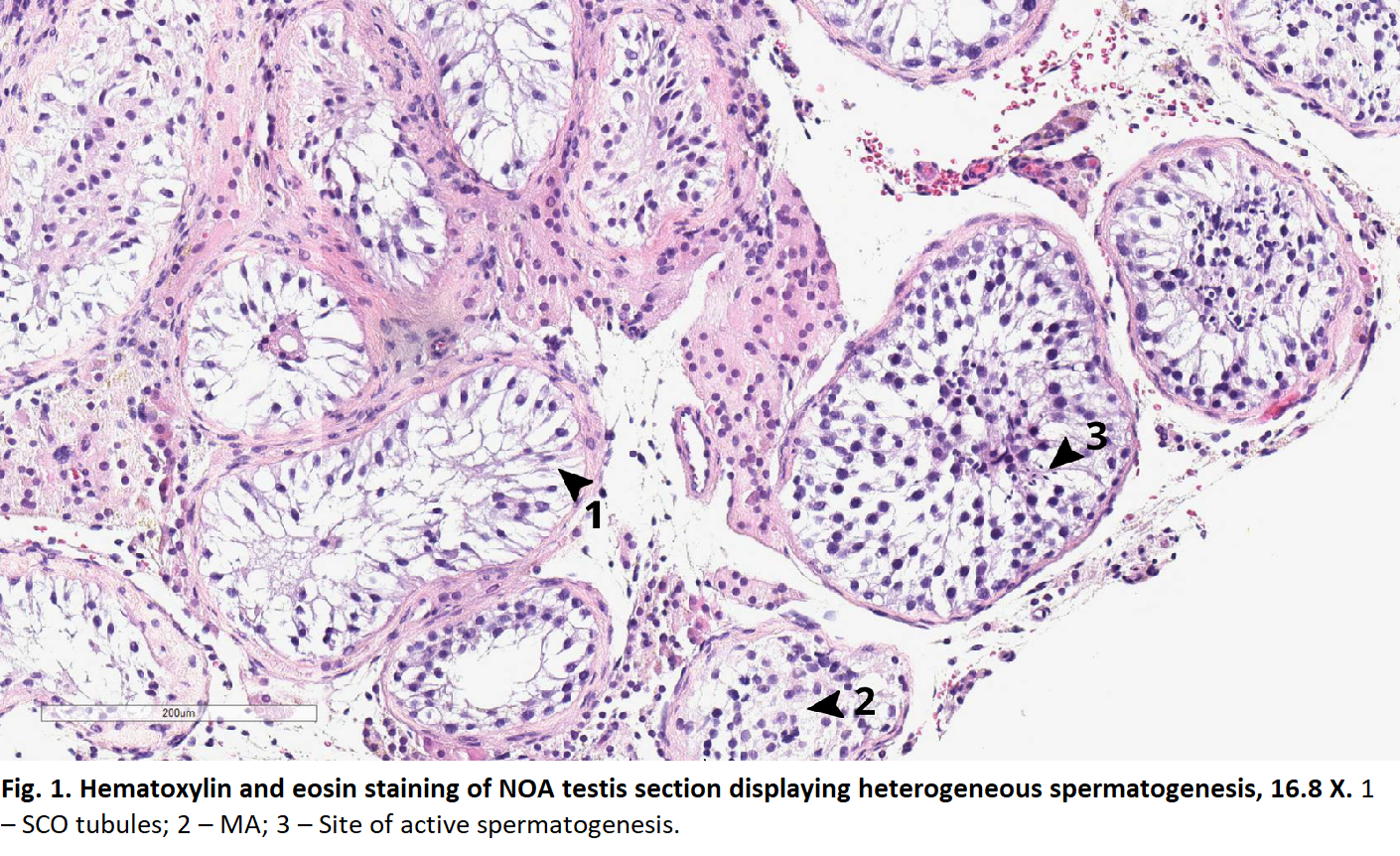
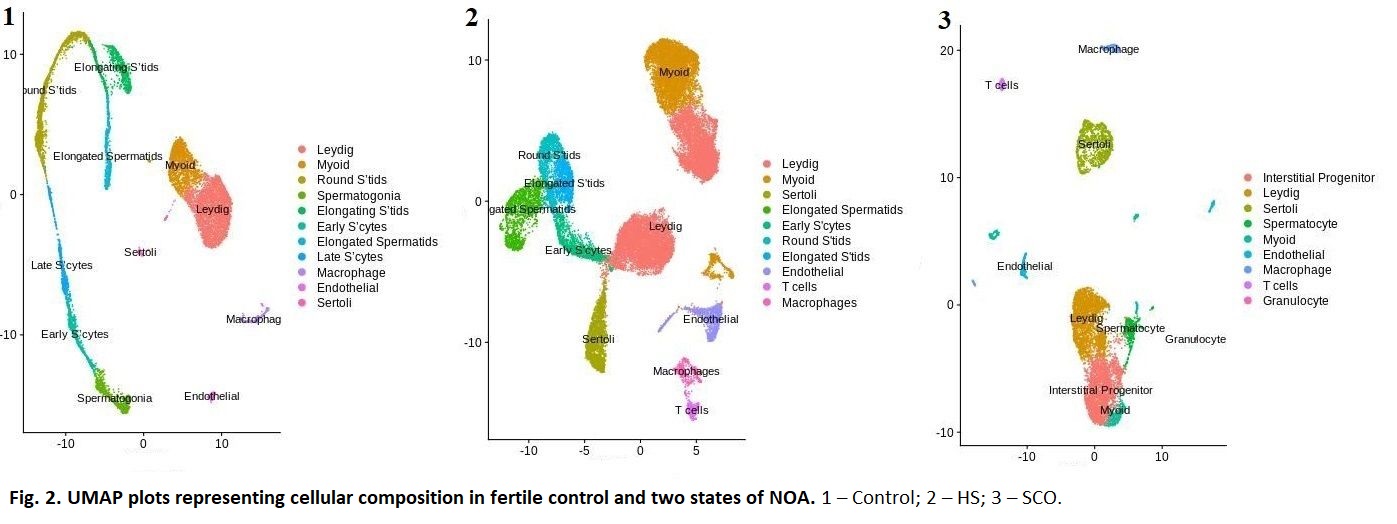
Methods: Two testis biopsies were derived from one patient with NOA with known histologic heterogeneity, and one normal control. ScRNAseq was carried out using 10X Genomics and Illumina sequencing platforms in technical duplicates. Cell clustering, gene expression, Ingenuity Pathway Analysis (IPA), ligand-receptor analyses and TradeSeq pseudotime differential expression analyses were used to evaluate the datasets.
Results: Cell types detected in our normal control was congruent with the previously published dataset. One NOA sample largely lacked germ cells and was broadly categorized as Sertoli Cell Only (SCO). This sample uniquely demonstrated a large cluster of cells expressing a mixture of Leydig and myoid genes; this same cluster exhibited high expression of NR4A1–known marker of common progenitor of Leydig and myoid cells. T cells and granulocytes were also present while absent among normal controls. In the second NOA sample, we detected all germ cells up to elongated spermatids and labelled this as hypospermatogenesis (HS). In this HS NOA sample, T cells were also present. After aligning the normal germ cells and HS NOA germ cells in pseudo time, differential gene expression and IPA pathway analyses revealed that NOA-derived germ cells had down regulation in RNA and nucleotide base excision repair mechanisms suggesting intrinsic deficiencies. Histology corroborated our scRNAseq results.
Conclusions: Differentiating features of the SCO sample were the presence of T cells, granulocytes and an interstitial progenitor population. Pathway analyses of NOA germ cells suggest intrinsic abnormalities in RNA processing.
Source of Funding: Canadian Institutes of Health Research; American Society of Reproductive Medicine; Vancouver Coastal Health Research Institute



.jpg)
.jpg)