Back
Poster, Podium & Video Sessions
Podium
PD40: Uroradiology II
PD40-03: Digital pathology labels perform better than radiologist labels for training deep learning models to detect prostate cancer on MRI
Sunday, May 15, 2022
9:50 AM – 10:00 AM
Location: Room 252
Indrani Bhattacharya, David Lim, Christian Kunder, Han Lin Aung, Xingchen Liu, Wei Shao, Simon Soerensen, Richard Fan, Pejman Ghanouni, Katherine To'o, James Brooks, Mirabela Rusu, Geoffrey Sonn*, Palo Alto, CA
- GS
Geoffrey A. Sonn, MD
Associate Professor of Urology
Podium Presenter(s)
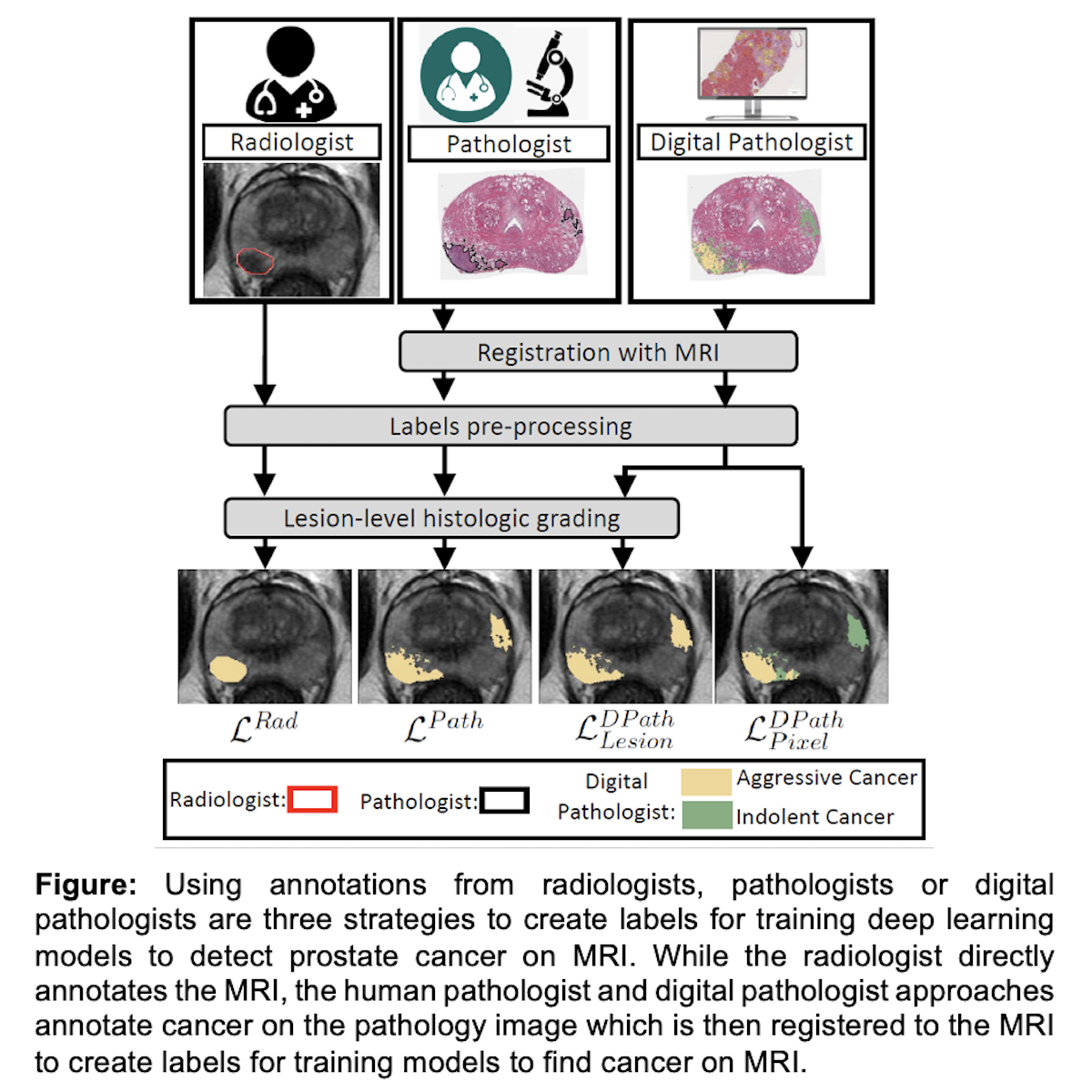
Introduction: Deep learning models to detect prostate cancer on Magnetic Resonance Imaging (MRI) have great potential to reduce inter-reader variability across radiologists. Our study compares different ground truth labeling strategies (Figure) to ascertain the best labels for training deep learning models to detect cancer on MRI.
Methods: We compared four kinds of ground truth labels in 115 patients who underwent radical prostatectomy: (1) radiologist labels confirmed to contain cancer, (2) human pathologist labels on whole-mount pathology images, (3) lesion-level digital pathologist labels from a validated deep learning model used on whole-mount pathology images and (4) pixel-level digital pathologist labels. Primary outcomes were lesion-level ROC AUC, lesion volume, and Dice coefficient.
Results: Radiologist labels have lower lesion-detection rates than pathology labels, and are not intended to capture the entire extent of cancer (lesion-level ROC-AUC: 0.75 - 0.84, lesion volumes ~75% of true cancer size on pathology, and Dice compared to pathology labels: 0.24 - 0.28). On a patient-level, 18/115 patients (16%) were false negatives on MRI, meaning that the cancer found on whole-mount pathology was not seen or annotated by the interpreting radiologist. In contrast, digital pathologist labels have high concordance with human pathologist labels (lesion ROC-AUC: 0.97 - 1, lesion Dice: 0.75 - 0.93). As a result, deep learning models trained using digital pathology labels performed better (ROC-AUC 0.91 - 0.94 for detection of Gleason =7 cancers) than models trained with radiologist labels. Moreover, pixel-level digital pathologist labels enable selective identification of aggressive and indolent cancer components in mixed lesions. This is not feasible for any human-annotated label type.
Conclusions: Labeling prostate MRI using information from whole mount pathology enables more accurate cancer annotation than relying upon labels from radiologists. Moreover, using digital pathology labels reduces challenges associated with human annotations, including time and inter- and intra-reader variability. Pathology-based strategies (human or digital) for MRI labeling improves the accuracy of deep learning models for cancer detection on MRI.
Source of Funding: Departments of Radiology and Urology, Stanford University, GE Healthcare Blue Sky Award
Methods: We compared four kinds of ground truth labels in 115 patients who underwent radical prostatectomy: (1) radiologist labels confirmed to contain cancer, (2) human pathologist labels on whole-mount pathology images, (3) lesion-level digital pathologist labels from a validated deep learning model used on whole-mount pathology images and (4) pixel-level digital pathologist labels. Primary outcomes were lesion-level ROC AUC, lesion volume, and Dice coefficient.
Results: Radiologist labels have lower lesion-detection rates than pathology labels, and are not intended to capture the entire extent of cancer (lesion-level ROC-AUC: 0.75 - 0.84, lesion volumes ~75% of true cancer size on pathology, and Dice compared to pathology labels: 0.24 - 0.28). On a patient-level, 18/115 patients (16%) were false negatives on MRI, meaning that the cancer found on whole-mount pathology was not seen or annotated by the interpreting radiologist. In contrast, digital pathologist labels have high concordance with human pathologist labels (lesion ROC-AUC: 0.97 - 1, lesion Dice: 0.75 - 0.93). As a result, deep learning models trained using digital pathology labels performed better (ROC-AUC 0.91 - 0.94 for detection of Gleason =7 cancers) than models trained with radiologist labels. Moreover, pixel-level digital pathologist labels enable selective identification of aggressive and indolent cancer components in mixed lesions. This is not feasible for any human-annotated label type.
Conclusions: Labeling prostate MRI using information from whole mount pathology enables more accurate cancer annotation than relying upon labels from radiologists. Moreover, using digital pathology labels reduces challenges associated with human annotations, including time and inter- and intra-reader variability. Pathology-based strategies (human or digital) for MRI labeling improves the accuracy of deep learning models for cancer detection on MRI.
Source of Funding: Departments of Radiology and Urology, Stanford University, GE Healthcare Blue Sky Award


.jpg)
.jpg)