Back
Poster Session C
Genetics, genomics and proteomics
Session: (1118–1149) Genetics, Genomics and Proteomics Poster
1126: Functional NOTCH4 Variants Increase Notch Signaling and Susceptibility for Systemic Sclerosis
Sunday, November 13, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- UK
Urvashi Kaundal, PhD
National Institutes of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), National Institutes of Health (NIH)
Chevy Chase, MD, United States
Abstract Poster Presenter(s)
Urvashi Kaundal1, Emilee Stenson1, Mousumi Sahu1, Krishan Kumar Thakur1, Janet Wang1, Ami Shah2, Maureen Mayes3, Ayo Doumatey4, Amy Bentley4, Daniel Shriner4, Robyn Domsic5, Thomas Medsger6, Paula Ramos7, Richard Silver7, Virginia Steen8, John Varga9, Vivien Hsu10, Lesley Ann Saketkoo11, Elena Schiopu12, Dinesh Khanna13, Jessica Gordon14, Lindsey Criswell15, Heather Gladue16, Chris Derk17, Elana Bernstein18, S. Louis Bridges, Jr.14, Victoria Shanmugam19, Lorinda Chung20, Suzanne Kafaja21, Reem Jan22, Marcin Trojanowski23, Avram Goldberg24, Benjamin Korman25, Jim Mullikin4, Stefania Dell'Orso1, Adebowale Adeyemo4, Charles Rotimi4, Elaine Remmers4, Daniel Kastner4, Fredrick Wigley26, Francesco Boin27 and Pravitt Gourh28, 1National Institutes of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), National Institutes of Health (NIH), Bethesda, MD, 2Johns Hopkins Rheumatology, Baltimore, MD, 3Division of Rheumatology and Clinical Immunogenetics, University of Texas McGovern Medical School, Houston, TX, 4National Human Genome Research Institute, Bethesda, MD, 5University of Pittsburgh, Pittsburgh, PA, 6University of Pittsburgh School of Medicine, Pittsburgh, PA, 7Medical University of South Carolina, Charleston, SC, 8Georgetown University School of Medicine, Washington, DC, 9University of Michigan, Ann Arbor, MI, 10Rutgers-RWJ Medical School, South Plainfield, NJ, 11University Medical Center - Comprehensive Pulmonary Hypertension Center and ILD Clinic Programs // New Orleans Scleroderma and Sarcoidosis Patient Care & Research Centeris, New Orleans, LA, 12Michigan Medicine, Ann Arbor, MI, 13Division of Rheumatology, Department of Internal Medicine, Scleroderma Program, University of Michigan, Ann Arbor, MI, 14Hospital for Special Surgery, New York, NY, 15National Human Genome Research Institute, NIH, Bethesda, MD, 16Arthritis & Osteoporosis Consultants of the Carolinas, Charlotte, NC, 17University of Pennsylvania, Philadelphia, PA, 18Columbia University, New York, NY, 19George Washington University, Great Falls, VA, 20Stanford University, Stanford, CA, 21UCLA Department of Medicine, Division of Rheumatology, Los Angeles, CA, 22University of Chicago, Chicago, IL, 23Boston University School of Medicine, Boston, MA, 24NYU Langone Medical Center - NYU Hospital for Joint Diseases, Lake Success, NY, 25University of Rochester, Rochester, NY, 26Johns Hopkins University, Baltimore, MD, 27Cedars-Sinai Medical Center, Los Angeles, CA, 28National Institutes of Health, Bethesda, MD
Background/Purpose: Genome wide association studies (GWAS) in systemic sclerosis (SSc) have identified several genetic loci, but the search for the causal variant and gene continues. In this study, using the Genome Research in African American Scleroderma Patients (GRASP) cohort, we evaluated previously reported genes from GWAS in SSc, primarily conducted in European ancestral populations. We further characterized the functional role of two variants in the Neurogenic Locus Notch Homolog Protein 4 (NOTCH4) gene.
Methods: We identified 32 genes associated with SSc susceptibility at p-value < 10-6 in the GWAS catalog. The gene-based sequence kernel association test (SKAT) test was used for association analysis and Bonferroni's correction for multiple testing. Expression quantitative trait loci (eQTL) analysis was performed using the Genotype-Tissue Expression (GTEx) data. Lymphoblastoid cell lines (LCLs) from the 1000 Genomes Project that were wildtype (WT) and heterozygous (HET) for the rs8192564 (a 5' UTR variant with CADD score >15) and rs17604492 (missense) were used to confirm the expression of NOTCH4 genes using RT-PCR and ELISA. Human lung microvascular endothelial cells (HULEC) were stimulated with NOTCH4 ligand-DLL4 to replicate increased signaling via this pathway.
Results: On comparing 379 AA SSc patients and 411 controls, only the NOTCH4 gene remained significant after multiple testing correction (Table 1A). This NOTCH4 association remained significant and was independent of the HLA variants, after conditional analysis. To validate these results, we examined an independent cohort of 590 patients and 360 controls and the NOTCH4 association remained significant. After multiple testing correction, diffuse cutaneous SSc, interstitial lung disease, and anti-fibrillarin antibody subsets remained statistically significant (Table 1B). eQTL analysis using GTEx data showed that both variants were associated with increased NOTCH4 expression in the HET LCLs and the rs17604492 variant also increased HEY2, (a downstream, nuclear signaling molecule for the Notch pathway) (Fig. 1B-D). Increased expression of NOTCH4 was confirmed in rs8192564 HET LCLs by RT-PCR (Fig. 1E). ELISA and western blot analysis using the lysates from LCLs confirmed the increased expression of NOTCH4 in LCLs carrying the risk alleles (Fig. 1F, G). Increased expression of the HEY2 and ACTA2 genes was observed on stimulating HULECs with DLL4 (Fig. 1 H, I). A previously published study has shown increased NOTCH4 expression in the skin of SSc patients (Fig. 2 A-D).
Conclusion: Functional variants in the NOTCH4 gene are associated with AA SSc patients and are independent of the HLA genes. Increased Notch signaling in endothelial cells can induce endothelial-to-mesenchymal transition and increased ACTA2 expression along with neoangiogenic circulation with sparse branching and dilated capillaries. SSc associated variants are associated with increased NOTCH4 expression and constitutively increase Notch4 signaling. Inhibitors of the gamma-secretase complex that cause cleavage of Notch intracellular domain have been shown to have a potent anti-fibrotic effect in different murine models of SSc and could be a potential therapeutic agent in SSc.
.jpg)
.jpg) Fig. 1. A. NOTCH4 rs17604492 variant alters glycine to arginine at position 942; B. rs8192564 eQTL analysis for wild type (WT) and heterozygous (Het) LCLs; C, D. rs17604492 eQTL analysis for WT and het LCLs; E. RT-PCR for NOTCH4 expression in LCLs WT and het for risk variants; F. ELISA for NOTCH4 protein in LCLs WT and het for risk variants; G. Western blot for NOTCH4 protein in LCLs and endothelial cells; H. RNA-seq for HEY2; I. RNA-seq for ACTA2. *normalized expression
Fig. 1. A. NOTCH4 rs17604492 variant alters glycine to arginine at position 942; B. rs8192564 eQTL analysis for wild type (WT) and heterozygous (Het) LCLs; C, D. rs17604492 eQTL analysis for WT and het LCLs; E. RT-PCR for NOTCH4 expression in LCLs WT and het for risk variants; F. ELISA for NOTCH4 protein in LCLs WT and het for risk variants; G. Western blot for NOTCH4 protein in LCLs and endothelial cells; H. RNA-seq for HEY2; I. RNA-seq for ACTA2. *normalized expression
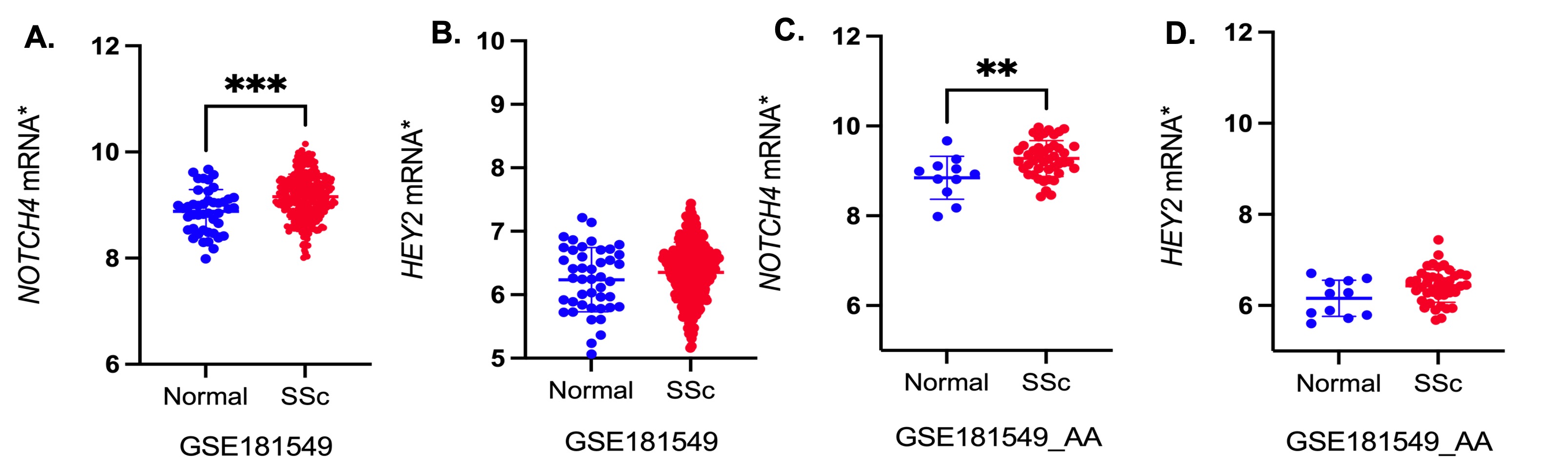 Fig. 2. A, B. Differential expression of NOTCH4 and HEY2 in SSc and normal skin; C, D. Differential expression of NOTCH4 and HEY2 in AA SSc and AA normal skin. *normalized expression.
Fig. 2. A, B. Differential expression of NOTCH4 and HEY2 in SSc and normal skin; C, D. Differential expression of NOTCH4 and HEY2 in AA SSc and AA normal skin. *normalized expression.
Disclosures: U. Kaundal, None; E. Stenson, None; M. Sahu, None; K. Thakur, None; J. Wang, None; A. Shah, Arena Pharmaceuticals, Medpace/Eicos, Kadmon Corporation; M. Mayes, Actelion Pharma, Mitsubishi-Tanabe, Boehringer Ingelheim, EICOS, Horizon Pharma, Prometheus, Corbus, Medtelligence; A. Doumatey, None; A. Bentley, None; D. Shriner, None; R. Domsic, None; T. Medsger, None; P. Ramos, None; R. Silver, None; V. Steen, None; J. Varga, Boehringer-Ingelheim; V. Hsu, None; L. Saketkoo, None; E. Schiopu, None; D. Khanna, Boehringer Ingelheim, Genentech, Prometheus, Horizon, Chemomab, Talaris, Gesynta, Amgen, Acceleron, Actelion, Bayer, CSL Behring, Paracrine Cell Therapy, Mitsubishi Tanabe, Theraly, Eicos Sciences; J. Gordon, None; L. Criswell, None; H. Gladue, GlaxoSmithKlein(GSK), AstraZeneca; C. Derk, None; E. Bernstein, Boehringer-Ingelheim, Kadmon, Pfizer; S. Bridges, Jr., Bristol Myers Squibb; V. Shanmugam, None; L. Chung, Kyverna, Mitsubishi Tanabe, Eicos, Boehringer-Ingelheim, Jasper, Genentech; S. Kafaja, None; R. Jan, None; M. Trojanowski, None; A. Goldberg, None; B. Korman, None; J. Mullikin, None; S. Dell'Orso, None; A. Adeyemo, None; C. Rotimi, None; E. Remmers, None; D. Kastner, None; F. Wigley, None; F. Boin, None; P. Gourh, None.
Background/Purpose: Genome wide association studies (GWAS) in systemic sclerosis (SSc) have identified several genetic loci, but the search for the causal variant and gene continues. In this study, using the Genome Research in African American Scleroderma Patients (GRASP) cohort, we evaluated previously reported genes from GWAS in SSc, primarily conducted in European ancestral populations. We further characterized the functional role of two variants in the Neurogenic Locus Notch Homolog Protein 4 (NOTCH4) gene.
Methods: We identified 32 genes associated with SSc susceptibility at p-value < 10-6 in the GWAS catalog. The gene-based sequence kernel association test (SKAT) test was used for association analysis and Bonferroni's correction for multiple testing. Expression quantitative trait loci (eQTL) analysis was performed using the Genotype-Tissue Expression (GTEx) data. Lymphoblastoid cell lines (LCLs) from the 1000 Genomes Project that were wildtype (WT) and heterozygous (HET) for the rs8192564 (a 5' UTR variant with CADD score >15) and rs17604492 (missense) were used to confirm the expression of NOTCH4 genes using RT-PCR and ELISA. Human lung microvascular endothelial cells (HULEC) were stimulated with NOTCH4 ligand-DLL4 to replicate increased signaling via this pathway.
Results: On comparing 379 AA SSc patients and 411 controls, only the NOTCH4 gene remained significant after multiple testing correction (Table 1A). This NOTCH4 association remained significant and was independent of the HLA variants, after conditional analysis. To validate these results, we examined an independent cohort of 590 patients and 360 controls and the NOTCH4 association remained significant. After multiple testing correction, diffuse cutaneous SSc, interstitial lung disease, and anti-fibrillarin antibody subsets remained statistically significant (Table 1B). eQTL analysis using GTEx data showed that both variants were associated with increased NOTCH4 expression in the HET LCLs and the rs17604492 variant also increased HEY2, (a downstream, nuclear signaling molecule for the Notch pathway) (Fig. 1B-D). Increased expression of NOTCH4 was confirmed in rs8192564 HET LCLs by RT-PCR (Fig. 1E). ELISA and western blot analysis using the lysates from LCLs confirmed the increased expression of NOTCH4 in LCLs carrying the risk alleles (Fig. 1F, G). Increased expression of the HEY2 and ACTA2 genes was observed on stimulating HULECs with DLL4 (Fig. 1 H, I). A previously published study has shown increased NOTCH4 expression in the skin of SSc patients (Fig. 2 A-D).
Conclusion: Functional variants in the NOTCH4 gene are associated with AA SSc patients and are independent of the HLA genes. Increased Notch signaling in endothelial cells can induce endothelial-to-mesenchymal transition and increased ACTA2 expression along with neoangiogenic circulation with sparse branching and dilated capillaries. SSc associated variants are associated with increased NOTCH4 expression and constitutively increase Notch4 signaling. Inhibitors of the gamma-secretase complex that cause cleavage of Notch intracellular domain have been shown to have a potent anti-fibrotic effect in different murine models of SSc and could be a potential therapeutic agent in SSc.
.jpg)
.jpg) Fig. 1. A. NOTCH4 rs17604492 variant alters glycine to arginine at position 942; B. rs8192564 eQTL analysis for wild type (WT) and heterozygous (Het) LCLs; C, D. rs17604492 eQTL analysis for WT and het LCLs; E. RT-PCR for NOTCH4 expression in LCLs WT and het for risk variants; F. ELISA for NOTCH4 protein in LCLs WT and het for risk variants; G. Western blot for NOTCH4 protein in LCLs and endothelial cells; H. RNA-seq for HEY2; I. RNA-seq for ACTA2. *normalized expression
Fig. 1. A. NOTCH4 rs17604492 variant alters glycine to arginine at position 942; B. rs8192564 eQTL analysis for wild type (WT) and heterozygous (Het) LCLs; C, D. rs17604492 eQTL analysis for WT and het LCLs; E. RT-PCR for NOTCH4 expression in LCLs WT and het for risk variants; F. ELISA for NOTCH4 protein in LCLs WT and het for risk variants; G. Western blot for NOTCH4 protein in LCLs and endothelial cells; H. RNA-seq for HEY2; I. RNA-seq for ACTA2. *normalized expression Fig. 2. A, B. Differential expression of NOTCH4 and HEY2 in SSc and normal skin; C, D. Differential expression of NOTCH4 and HEY2 in AA SSc and AA normal skin. *normalized expression.
Fig. 2. A, B. Differential expression of NOTCH4 and HEY2 in SSc and normal skin; C, D. Differential expression of NOTCH4 and HEY2 in AA SSc and AA normal skin. *normalized expression.Disclosures: U. Kaundal, None; E. Stenson, None; M. Sahu, None; K. Thakur, None; J. Wang, None; A. Shah, Arena Pharmaceuticals, Medpace/Eicos, Kadmon Corporation; M. Mayes, Actelion Pharma, Mitsubishi-Tanabe, Boehringer Ingelheim, EICOS, Horizon Pharma, Prometheus, Corbus, Medtelligence; A. Doumatey, None; A. Bentley, None; D. Shriner, None; R. Domsic, None; T. Medsger, None; P. Ramos, None; R. Silver, None; V. Steen, None; J. Varga, Boehringer-Ingelheim; V. Hsu, None; L. Saketkoo, None; E. Schiopu, None; D. Khanna, Boehringer Ingelheim, Genentech, Prometheus, Horizon, Chemomab, Talaris, Gesynta, Amgen, Acceleron, Actelion, Bayer, CSL Behring, Paracrine Cell Therapy, Mitsubishi Tanabe, Theraly, Eicos Sciences; J. Gordon, None; L. Criswell, None; H. Gladue, GlaxoSmithKlein(GSK), AstraZeneca; C. Derk, None; E. Bernstein, Boehringer-Ingelheim, Kadmon, Pfizer; S. Bridges, Jr., Bristol Myers Squibb; V. Shanmugam, None; L. Chung, Kyverna, Mitsubishi Tanabe, Eicos, Boehringer-Ingelheim, Jasper, Genentech; S. Kafaja, None; R. Jan, None; M. Trojanowski, None; A. Goldberg, None; B. Korman, None; J. Mullikin, None; S. Dell'Orso, None; A. Adeyemo, None; C. Rotimi, None; E. Remmers, None; D. Kastner, None; F. Wigley, None; F. Boin, None; P. Gourh, None.

