Back
Poster Session C
Rheumatoid arthritis (RA)
Session: (1417–1439) RA – Treatment Poster III
1428: Molecular and Clinical Profiling of Rheumatoid Arthritis Patients Predicts the Response to Rituximab
Sunday, November 13, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- DT
Daniel Toro-Domínguez, Sr, PhD
GENYO
Granada, Spain
Abstract Poster Presenter(s)
Daniel Toro-Domínguez1, José Linares-Blanco1, Raúl López-Domínguez2, Pilar S. López-Garrido2, Georgina Galicia-Rosas1, Pedro Carmona-Sáez2 and Marta Alarcon-Riquelme1, 1Center for Genomics and Oncological Research (GENYO), Granada, Spain, 2University of Granada, Granada, Spain
Background/Purpose: Rheumatoid arthritis (RA) is one of the most prevalent rheumatic diseases, mainly characterized by chronically painful, swollen joints that can severely impair physical function and quality of life. Patients can present a wide variety of clinical manifestations and therapeutic approaches are only partially effective. There is a knowledge-gap in the molecular mechanisms behind response or non-response to the treatments, which are often applied in a trial-and-error approach. This not only entails an economic cost, but disease continues to progress in those patients who do not respond to the applied therapy. Therefore, advances in the knowledge of the molecular and clinical patterns that condition the response, as well as the development of new guidelines or tools with which to predict response in a personalized way are urgently needed.
Methods: Machine learning models to predict clinical response to rituximab based on gene expression data was created in a dataset with 23 and 24 responder and non-responder RA patients, respectively. First, 80 and 20 percent of patients were randomly selected as training and test set. Different algorithms were tested. To select optimal parameter values, parameter tuning was performed on the training set for each model by 10-fold-cross validation. Performance results of each algorithm were measured on the test set. The entire process was repeated 10 times from train/test selection to avoid over-fitting and finally, the best model was selected based on the mean of the area under the curve (AUC) values across iterations. An independent cohort of RA patients was used to test model reproducibility. The model was used to predict response and non-response in a third cohort of 295 RA patients, which contained data on gene expression, methylation, metagenomics, and a large set of clinical information, including both clinical manifestations, autoantibodies, various cytokines and blood cell proportions. All of these types of data were analyzed comparing responders and non-responders to perform a multilayered characterization of the molecular mechanisms behind drug response. A summary of the methods is provided in Figure 1.
Results: A prediction model with an AUC of 0.82 was obtained. A set of differentially expressed and methylated genes different between responders and non-responders was observed, mainly related to the activation and differentiation of T cells, as well as signaling by cytokines IL2 and IL7. No significant difference was found at the microbiome level, but significant differences in the proportions of different blood cells, including T cells, NK cells and low-density granulocytes, as well as differences at the level of comorbidities were found, being more frequent the presence of type II diabetes or dyslipidemia in non-responding patients.
Conclusion: A tool capable of classifying responder and non-responder patients was developed, which also provided novel information on the molecular basis behind rituximab response and non-response.
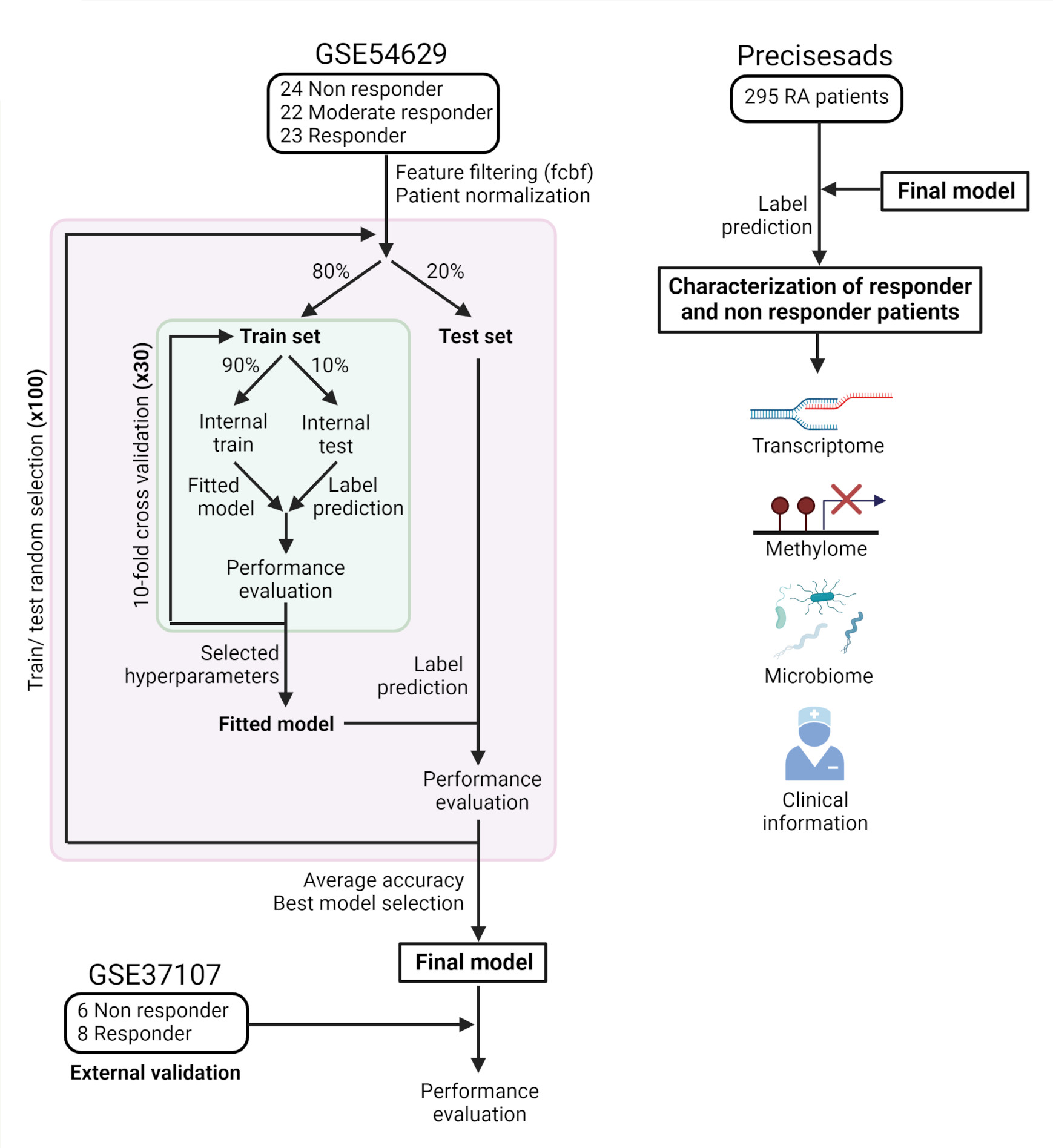 Figure 1: Summary diagram. The figure shows a diagram about the process of building the prediction model and the subsequent omic characterization of responders and non-responders patients.
Figure 1: Summary diagram. The figure shows a diagram about the process of building the prediction model and the subsequent omic characterization of responders and non-responders patients.
Disclosures: D. Toro-Domínguez, None; J. Linares-Blanco, None; R. López-Domínguez, None; P. López-Garrido, None; G. Galicia-Rosas, None; P. Carmona-Sáez, None; M. Alarcon-Riquelme, None.
Background/Purpose: Rheumatoid arthritis (RA) is one of the most prevalent rheumatic diseases, mainly characterized by chronically painful, swollen joints that can severely impair physical function and quality of life. Patients can present a wide variety of clinical manifestations and therapeutic approaches are only partially effective. There is a knowledge-gap in the molecular mechanisms behind response or non-response to the treatments, which are often applied in a trial-and-error approach. This not only entails an economic cost, but disease continues to progress in those patients who do not respond to the applied therapy. Therefore, advances in the knowledge of the molecular and clinical patterns that condition the response, as well as the development of new guidelines or tools with which to predict response in a personalized way are urgently needed.
Methods: Machine learning models to predict clinical response to rituximab based on gene expression data was created in a dataset with 23 and 24 responder and non-responder RA patients, respectively. First, 80 and 20 percent of patients were randomly selected as training and test set. Different algorithms were tested. To select optimal parameter values, parameter tuning was performed on the training set for each model by 10-fold-cross validation. Performance results of each algorithm were measured on the test set. The entire process was repeated 10 times from train/test selection to avoid over-fitting and finally, the best model was selected based on the mean of the area under the curve (AUC) values across iterations. An independent cohort of RA patients was used to test model reproducibility. The model was used to predict response and non-response in a third cohort of 295 RA patients, which contained data on gene expression, methylation, metagenomics, and a large set of clinical information, including both clinical manifestations, autoantibodies, various cytokines and blood cell proportions. All of these types of data were analyzed comparing responders and non-responders to perform a multilayered characterization of the molecular mechanisms behind drug response. A summary of the methods is provided in Figure 1.
Results: A prediction model with an AUC of 0.82 was obtained. A set of differentially expressed and methylated genes different between responders and non-responders was observed, mainly related to the activation and differentiation of T cells, as well as signaling by cytokines IL2 and IL7. No significant difference was found at the microbiome level, but significant differences in the proportions of different blood cells, including T cells, NK cells and low-density granulocytes, as well as differences at the level of comorbidities were found, being more frequent the presence of type II diabetes or dyslipidemia in non-responding patients.
Conclusion: A tool capable of classifying responder and non-responder patients was developed, which also provided novel information on the molecular basis behind rituximab response and non-response.
 Figure 1: Summary diagram. The figure shows a diagram about the process of building the prediction model and the subsequent omic characterization of responders and non-responders patients.
Figure 1: Summary diagram. The figure shows a diagram about the process of building the prediction model and the subsequent omic characterization of responders and non-responders patients.Disclosures: D. Toro-Domínguez, None; J. Linares-Blanco, None; R. López-Domínguez, None; P. López-Garrido, None; G. Galicia-Rosas, None; P. Carmona-Sáez, None; M. Alarcon-Riquelme, None.

