Back
Poster Session C
Fibrosing rheumatic diseases (scleroderma, MCTD, IgG4-related disease, scleroderma mimics)
Session: (1166–1185) Systemic Sclerosis and Related Disorders – Basic Science Poster
1179: Single Cell RNA Sequencing Analysis of Cryopreserved versus Fresh Skin Biopsy Samples from Systemic Sclerosis Patients and Healthy Controls
Sunday, November 13, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
.jpg)
Brian Skaug, MD, PhD
University of Texas McGovern Medical School at Houston
Houston, TX, United States
Abstract Poster Presenter(s)
Brian Skaug1, Marka Lyons1, Julio Charles1, Maureen Mayes2 and Shervin Assassi3, 1University of Texas McGovern Medical School at Houston, Houston, TX, 2Division of Rheumatology and Clinical Immunogenetics, University of Texas McGovern Medical School, Houston, TX, 3McGovern Medical School, University of Texas, Houston, TX
Background/Purpose: Single cell RNA Sequencing (scRNA-Seq) can elucidate tissue-resident cell populations and gene expression profiles with unprecedented granularity. Application of this approach to systemic sclerosis (SSc) skin samples has begun to reveal novel insights into SSc pathogenesis. Cryopreservation of skin samples could facilitate this line of research by allowing batching and shipment of samples to centers with the requisite infrastructure to perform scRNA-Seq. However, it is important first to establish whether or not skin samples can be cryopreserved without compromising their quality and substantively affecting the results from scRNA-Seq analyses. To address this question, we compared scRNA-Seq results from paired skin biopsy samples in which one sample was freshly prepared and the other was cryopreserved prior to single cell isolation and analysis.
Methods: Three diffuse cutaneous SSc patients and two healthy controls underwent paired forearm skin biopsies. One sample from each subject was immediately subjected to dissociation, followed by single cell capture using the 10X Genomics Chromium Controller. The other sample was placed in CryoStor CS10 solution and stored in liquid nitrogen for 2-5 weeks, then thawed and subjected to the same protocol for dissociation and single cell capture. Roughly 2,000 nucleated cells per sample were subjected to single cell capture and analysis. 10X Genomics 3' gene expression protocol was followed for preparation of sequencing libraries. Each sample was sequenced at a depth of roughly 100 million reads (50,000 reads/cell). Sequencing results were analyzed for quality metrics including reads and genes per cell and percent of reads confidently mapped to exonic regions. Cell clustering, cell type and gene expression analyses were performed using the Seurat package in R.
Results: Cell viability was lower in cryopreserved compared to fresh samples (median difference 13%, range 4 – 41%) (Fig. 1A). Reads/cell, genes/cell, and percent of reads confidently mapped to exonic regions were lower in cryopreserved compared to fresh samples (Fig. 1B-D). When analyzed together, there was only partial overlap in gene expression between fresh and cryopreserved samples (Fig. 2A). Some of the differences observed in cryopreserved samples corresponded to areas with low detected genes/cell (Fig. 2B). Analyzed separately, there were differences in cell clusters identified in cryopreserved compared to fresh samples (Fig. 3A). Some cell populations, for example CD3-positive cells, appeared unaffected by cryopreservation (Fig. 3B). By contrast, innate immune cells were affected, with a reduction in CCL22-positive and CD163-positive cell counts and loss of resolution between these two cell populations in cryopreserved samples (Fig. 3C).
Conclusion: Cryopreservation of skin samples from SSc patients and healthy controls caused reduced cell viability, reduced quality of scRNA-Seq information, and alterations in some of the detected cell populations and gene expression profiles. Our results suggest potential drawbacks of utilizing cryopreserved skin samples for scRNA-Seq studies of SSc patients and healthy controls.
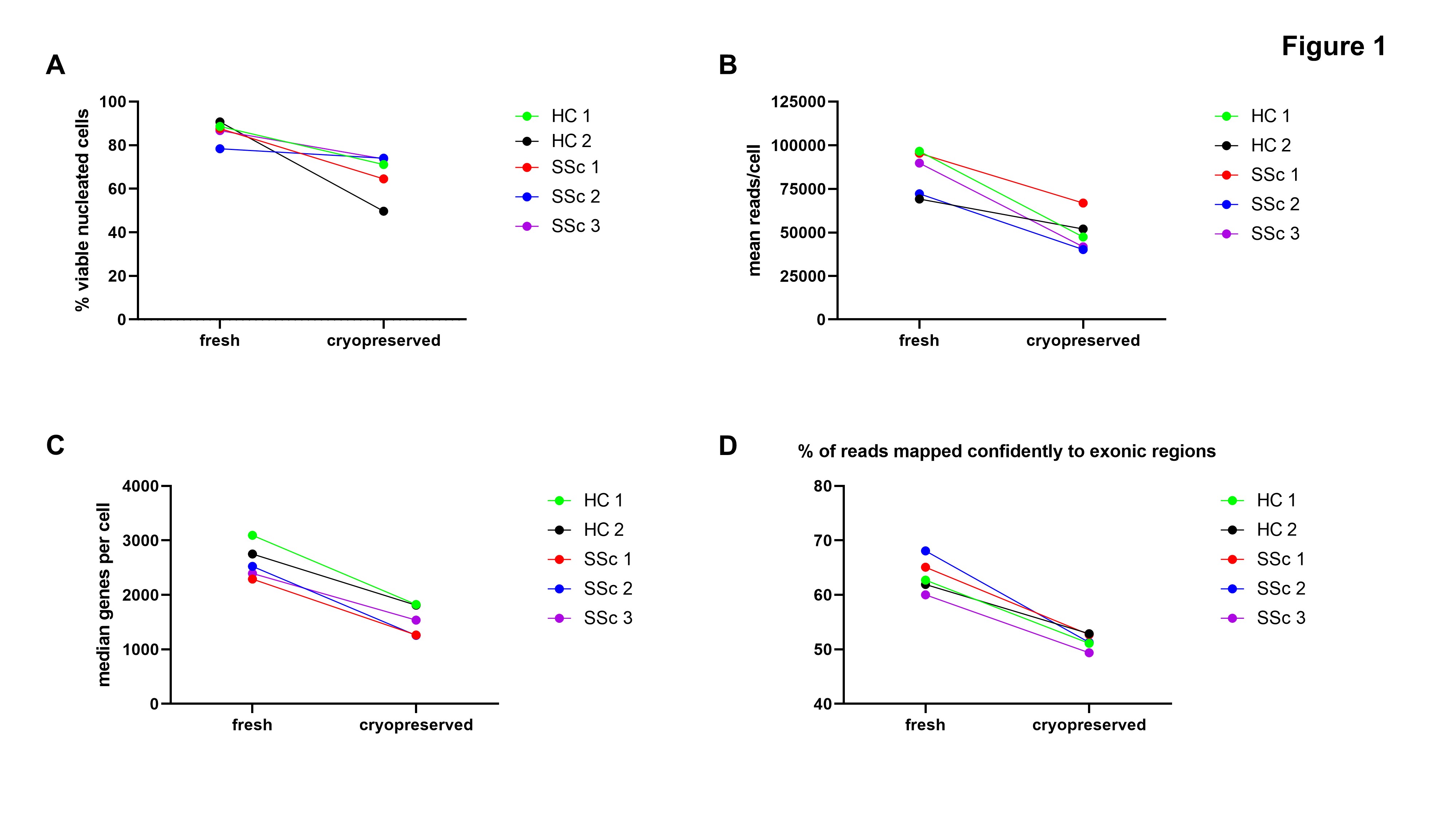 Figure 1: (A) Viability of nucleated cells from fresh vs. cryopreserved skin samples after skin dissociation. (B-D) Quality metrics after single cell RNA Sequencing of fresh vs. cryopreserved samples. HC: healthy control, SSc: systemic sclerosis
Figure 1: (A) Viability of nucleated cells from fresh vs. cryopreserved skin samples after skin dissociation. (B-D) Quality metrics after single cell RNA Sequencing of fresh vs. cryopreserved samples. HC: healthy control, SSc: systemic sclerosis
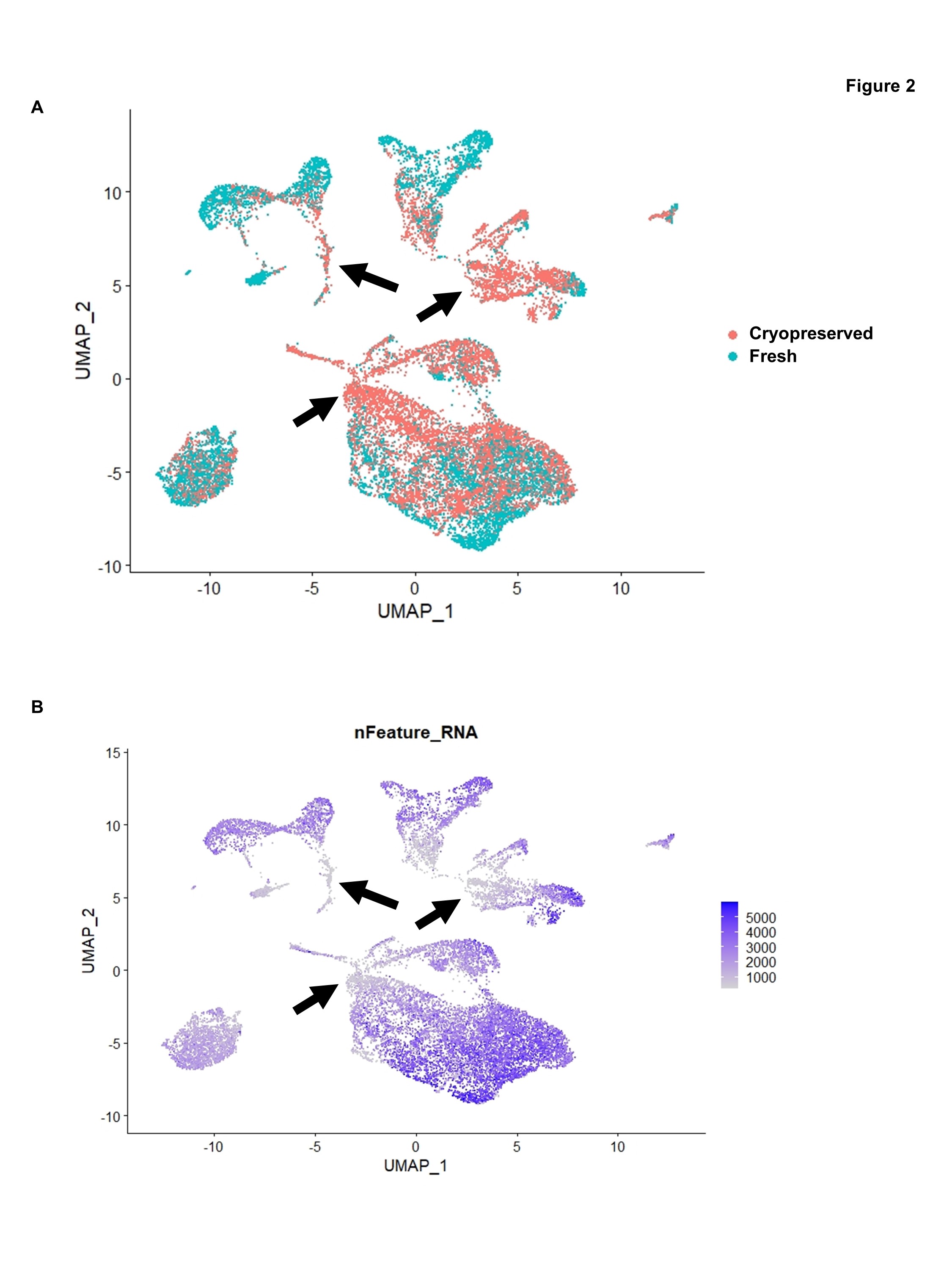 Figure 2: (A) UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples. (B) Same as (A), but highlighting nFeature RNA, which corresponds to the number of genes per cell. Black arrows indicate groups of cells with low nFeature RNA that are made up predominantly of cells from cryopreserved samples.
Figure 2: (A) UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples. (B) Same as (A), but highlighting nFeature RNA, which corresponds to the number of genes per cell. Black arrows indicate groups of cells with low nFeature RNA that are made up predominantly of cells from cryopreserved samples.
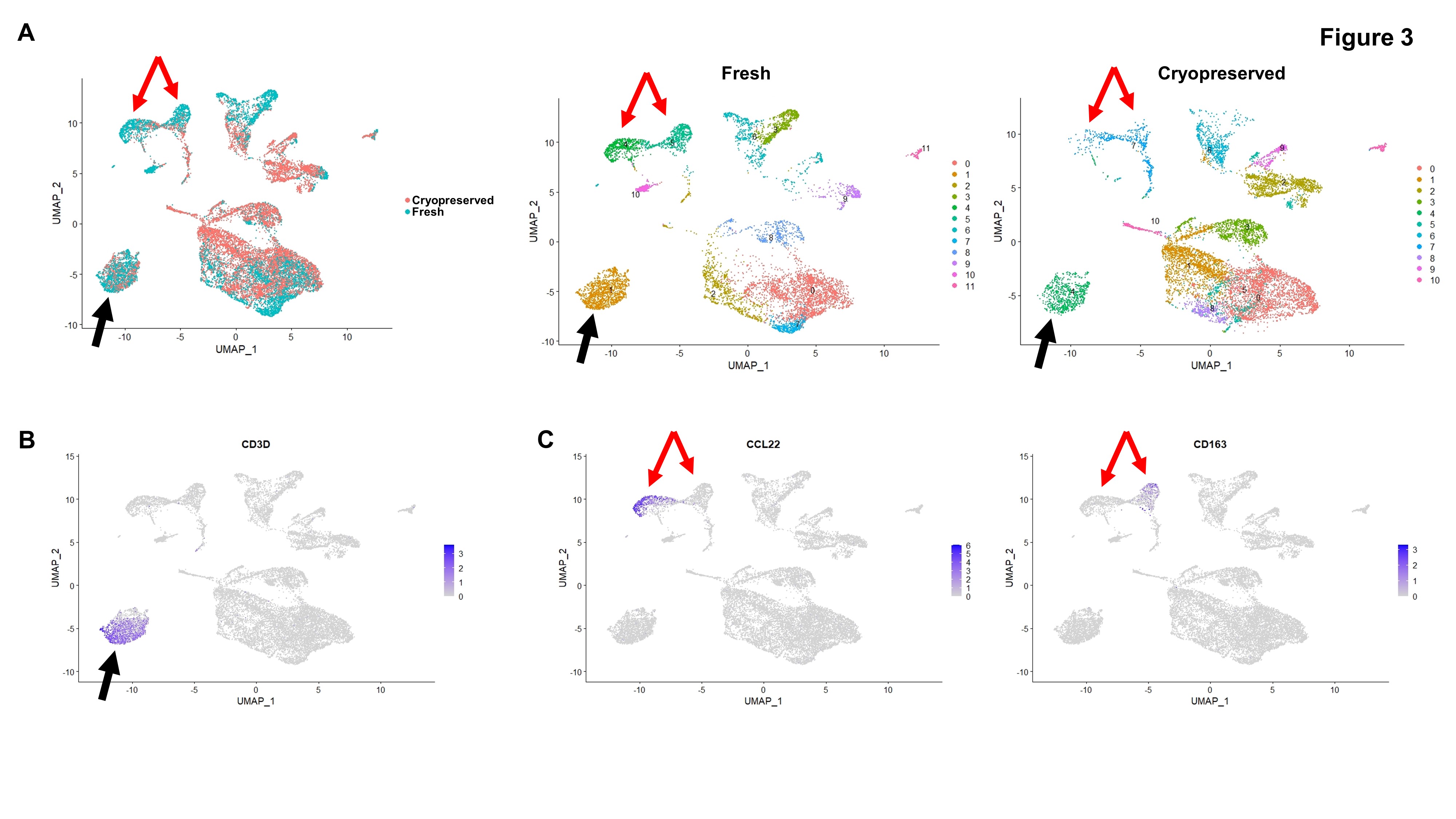 Figure 3: (A) Left: UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples (Same as Figure 2A). Right: Clusters of cells identified based on gene expression profiles in fresh vs. cryopreserved samples. Black and red arrows indicate areas that are highlighted in Figure 3B (black arrow) and 3C (red arrows). (B) Feature plot showing CD3-expressing cells. (C) Feature plots showing CCL22- and CD163-expressing cells.
Figure 3: (A) Left: UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples (Same as Figure 2A). Right: Clusters of cells identified based on gene expression profiles in fresh vs. cryopreserved samples. Black and red arrows indicate areas that are highlighted in Figure 3B (black arrow) and 3C (red arrows). (B) Feature plot showing CD3-expressing cells. (C) Feature plots showing CCL22- and CD163-expressing cells.
Disclosures: B. Skaug, None; M. Lyons, None; J. Charles, None; M. Mayes, Actelion Pharma, Mitsubishi-Tanabe, Boehringer Ingelheim, EICOS, Horizon Pharma, Prometheus, Corbus, Medtelligence; S. Assassi, Boehringer-Ingelheim, Janssen, Novartis, AstraZeneca, CSL Behring, AbbVie/Abbott.
Background/Purpose: Single cell RNA Sequencing (scRNA-Seq) can elucidate tissue-resident cell populations and gene expression profiles with unprecedented granularity. Application of this approach to systemic sclerosis (SSc) skin samples has begun to reveal novel insights into SSc pathogenesis. Cryopreservation of skin samples could facilitate this line of research by allowing batching and shipment of samples to centers with the requisite infrastructure to perform scRNA-Seq. However, it is important first to establish whether or not skin samples can be cryopreserved without compromising their quality and substantively affecting the results from scRNA-Seq analyses. To address this question, we compared scRNA-Seq results from paired skin biopsy samples in which one sample was freshly prepared and the other was cryopreserved prior to single cell isolation and analysis.
Methods: Three diffuse cutaneous SSc patients and two healthy controls underwent paired forearm skin biopsies. One sample from each subject was immediately subjected to dissociation, followed by single cell capture using the 10X Genomics Chromium Controller. The other sample was placed in CryoStor CS10 solution and stored in liquid nitrogen for 2-5 weeks, then thawed and subjected to the same protocol for dissociation and single cell capture. Roughly 2,000 nucleated cells per sample were subjected to single cell capture and analysis. 10X Genomics 3' gene expression protocol was followed for preparation of sequencing libraries. Each sample was sequenced at a depth of roughly 100 million reads (50,000 reads/cell). Sequencing results were analyzed for quality metrics including reads and genes per cell and percent of reads confidently mapped to exonic regions. Cell clustering, cell type and gene expression analyses were performed using the Seurat package in R.
Results: Cell viability was lower in cryopreserved compared to fresh samples (median difference 13%, range 4 – 41%) (Fig. 1A). Reads/cell, genes/cell, and percent of reads confidently mapped to exonic regions were lower in cryopreserved compared to fresh samples (Fig. 1B-D). When analyzed together, there was only partial overlap in gene expression between fresh and cryopreserved samples (Fig. 2A). Some of the differences observed in cryopreserved samples corresponded to areas with low detected genes/cell (Fig. 2B). Analyzed separately, there were differences in cell clusters identified in cryopreserved compared to fresh samples (Fig. 3A). Some cell populations, for example CD3-positive cells, appeared unaffected by cryopreservation (Fig. 3B). By contrast, innate immune cells were affected, with a reduction in CCL22-positive and CD163-positive cell counts and loss of resolution between these two cell populations in cryopreserved samples (Fig. 3C).
Conclusion: Cryopreservation of skin samples from SSc patients and healthy controls caused reduced cell viability, reduced quality of scRNA-Seq information, and alterations in some of the detected cell populations and gene expression profiles. Our results suggest potential drawbacks of utilizing cryopreserved skin samples for scRNA-Seq studies of SSc patients and healthy controls.
 Figure 1: (A) Viability of nucleated cells from fresh vs. cryopreserved skin samples after skin dissociation. (B-D) Quality metrics after single cell RNA Sequencing of fresh vs. cryopreserved samples. HC: healthy control, SSc: systemic sclerosis
Figure 1: (A) Viability of nucleated cells from fresh vs. cryopreserved skin samples after skin dissociation. (B-D) Quality metrics after single cell RNA Sequencing of fresh vs. cryopreserved samples. HC: healthy control, SSc: systemic sclerosis Figure 2: (A) UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples. (B) Same as (A), but highlighting nFeature RNA, which corresponds to the number of genes per cell. Black arrows indicate groups of cells with low nFeature RNA that are made up predominantly of cells from cryopreserved samples.
Figure 2: (A) UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples. (B) Same as (A), but highlighting nFeature RNA, which corresponds to the number of genes per cell. Black arrows indicate groups of cells with low nFeature RNA that are made up predominantly of cells from cryopreserved samples. Figure 3: (A) Left: UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples (Same as Figure 2A). Right: Clusters of cells identified based on gene expression profiles in fresh vs. cryopreserved samples. Black and red arrows indicate areas that are highlighted in Figure 3B (black arrow) and 3C (red arrows). (B) Feature plot showing CD3-expressing cells. (C) Feature plots showing CCL22- and CD163-expressing cells.
Figure 3: (A) Left: UMAP display of cells from all five biopsy pairs, highlighting cells from fresh vs. cryopreserved samples (Same as Figure 2A). Right: Clusters of cells identified based on gene expression profiles in fresh vs. cryopreserved samples. Black and red arrows indicate areas that are highlighted in Figure 3B (black arrow) and 3C (red arrows). (B) Feature plot showing CD3-expressing cells. (C) Feature plots showing CCL22- and CD163-expressing cells.Disclosures: B. Skaug, None; M. Lyons, None; J. Charles, None; M. Mayes, Actelion Pharma, Mitsubishi-Tanabe, Boehringer Ingelheim, EICOS, Horizon Pharma, Prometheus, Corbus, Medtelligence; S. Assassi, Boehringer-Ingelheim, Janssen, Novartis, AstraZeneca, CSL Behring, AbbVie/Abbott.

