Back
Poster Session A
Systemic lupus erythematosus (SLE)
Session: (0317–0342) SLE – Diagnosis, Manifestations, and Outcomes Poster I: Diagnosis
0342: Multi-Parametric Interrogation of the Systemic Lupus Erythematosus (SLE) Immunome Reveals Multiple Derangements
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- KN
Katherine Nay Yaung, -None-
Duke-NUS Medical School
Singapore, Singapore
Abstract Poster Presenter(s)
Katherine Nay Yaung1, Joo Guan Yeo2, Hui Nee Annie Law3, Martin Wasser4, Thaschawee Arkachaisri5, Julian Thumboo3, Andrea Low3 and Salvatore Albani4, 1Translational Immunology Institute, SingHealth Duke-NUS Academic Medical Centre, Singapore; Duke-NUS Medical School, Singapore, Singapore, 2KK Hospital, Singapore; Translational Immunology Institute, SingHealth Duke-NUS Academic Medical Centre, Singapore, Singapore, 3Department of Rheumatology and Immunology, Singapore General Hospital, Singapore, Singapore, 4Translational Immunology Institute, SingHealth Duke-NUS Academic Medical Centre, Singapore, Singapore, Singapore, 5KK Women's and Children's Hospital, SingHealth, Singapore, Singapore
Background/Purpose: Systemic lupus erythematosus (SLE) is a complex, systemic autoimmune disease that interferes with the balance between regulation and immunity, resulting in immune system dysfunction. Disease course is unpredictable due to alternating remissions and flares, impeding reliable patient assessments. Thus, mechanistic insights are required for better assessment. Current studies are mainly descriptive, and SLE is best interrogated with a multi-parametric, holistic approach such as mass cytometry (CyTOF).
The objectives of this study are as follows: (1) To characterise immune signatures of SLE patients and in the process: Study the roles of B and T cells in SLE to gain a holistic understanding of the adaptive immune response and (2) To compare immunological profiles of SLE patients with age-matched healthy controls.
Methods: Peripheral blood mononuclear cells (PBMCs) from 26 SLE subjects (41 samples, median age 39.5years) and 27 age-matched subjects were tested with CyTOF. Data was analysed and visualised using our laboratory-designed machine learning tool, the Extended Polydimensional Immunome Characterization (EPIC) platform. Normalization and FlowSOM (Flow cytometry analysis by Self- Organising Maps) clustering were performed with functionally and phenotypically important immune markers. Mann-Whitney U test identified significantly different cluster frequencies.
Results: Multiple significant differences were identified (p < 0.05). An activated CD8+CXCR3+CLA+ T -cell subset (CD8+CD45RA+CD38+CLA+CXCR3+) was enriched in SLE (median: 0.19%,interquartile range: 0.13-0.35% of CD45+ PBMCs) versus healthy (0.08%,0.05-0.09%; p< 0.0001). CXCR3 and cutaneous lymphocyte-associated antigen (CLA) are involved in skin-homing, with CLA function enhanced in inflammatory dermatoses. This suggests the importance of skin involvement in SLE immunopathogenesis even if skin manifestations are absent. Further analysis of CD3+ cells revealed unchanged natural T-regulatory cells (TREGS) in healthy and disease while a few TREG-like populations (FoxP3+CD25-) are significantly increased in disease, suggesting a deranged immunoregulatory response driven by TREGS. Thirdly, an activated CD8+BAFF+ T-cell subset (CD8+CD45RA+iCOS+ BAFF+) (r=0.715, p< 0.0001) is enriched in SLE (0.975%,0.433-1.53%) versus healthy (0.28%,0.1-0.49%; p< 0.0001). B-cell activating factor (BAFF) supports autoreactive B cell survival in autoimmune disease; an anti-BAFF drug (belimumab) is FDA- approved for SLE. Despite increasing use of belimumab, not all patients respond equally, so BAFF inhibition alone may not adequately alter disease activity. Studying these cell subset interactions in further detail to identify pathological pathways may facilitate improvements in current SLE theragnostics.
Conclusion: With a multi-parametric unbiased approach comparing SLE subjects to healthy controls, we identified immune subsets of immunopathogenic importance for further studies.
 Figure 1: Clinical samples are used for experimentation and subsequent analysis.
Figure 1: Clinical samples are used for experimentation and subsequent analysis.
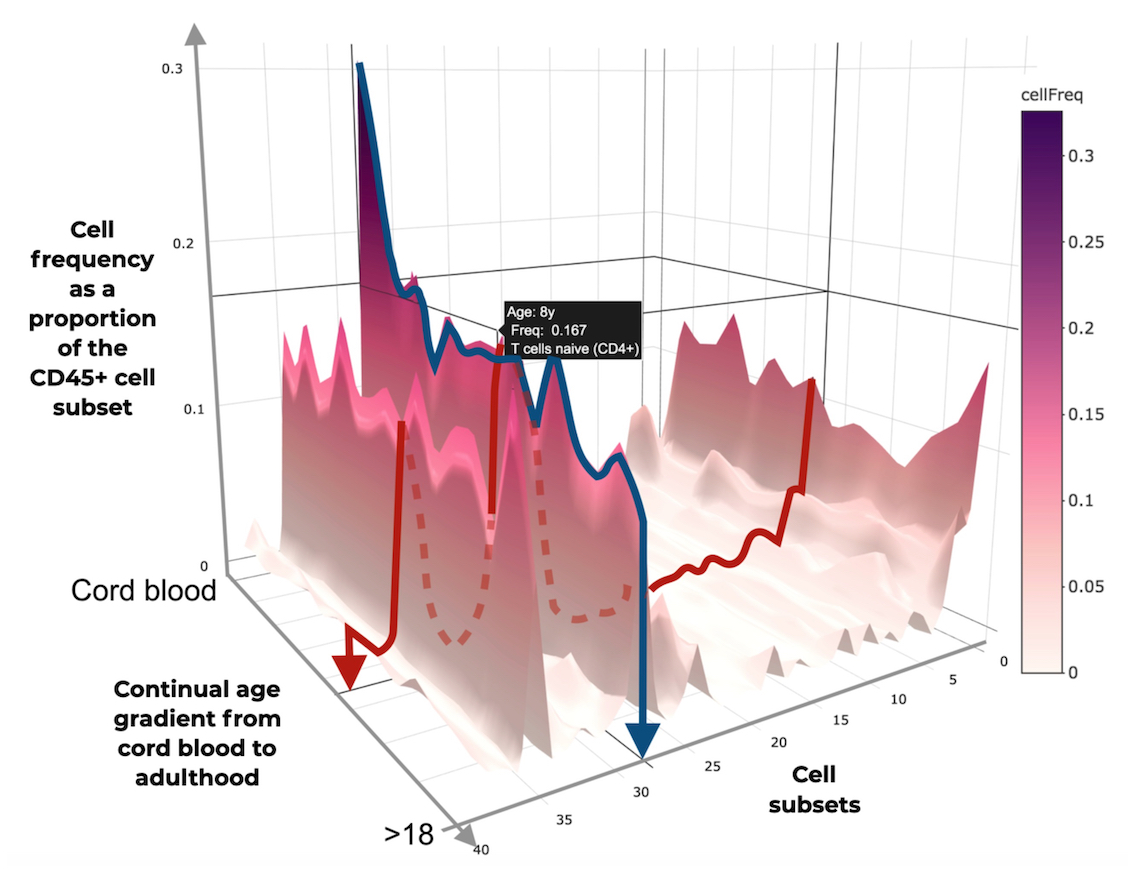 Figure 2: A graphic representation of the EPIC Immunomics Platform, which provides a healthy dataset and analysis pipeline to identify derangements in disease.
Figure 2: A graphic representation of the EPIC Immunomics Platform, which provides a healthy dataset and analysis pipeline to identify derangements in disease.
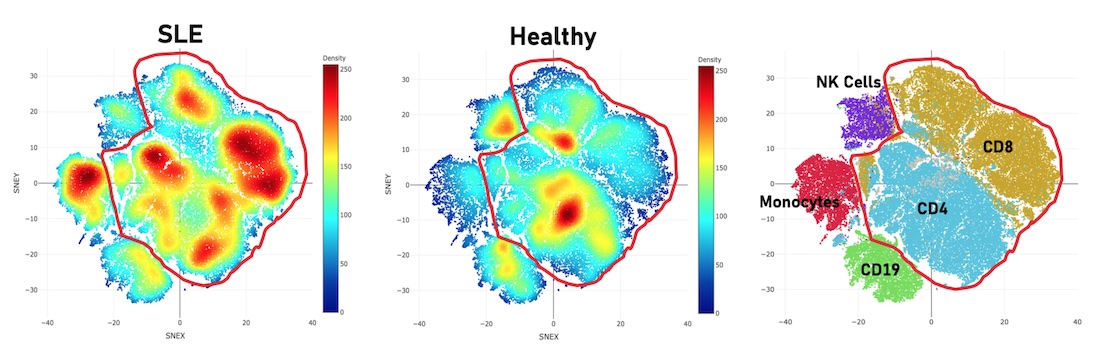 Figure 3: There are derangements in multiple subsets in SLE. Notably, many differences between SLE and healthy were observed in the CD3+ compartment (circled in red).
Figure 3: There are derangements in multiple subsets in SLE. Notably, many differences between SLE and healthy were observed in the CD3+ compartment (circled in red).
Disclosures: K. Nay Yaung, None; J. Yeo, None; H. Law, None; M. Wasser, None; T. Arkachaisri, None; J. Thumboo, None; A. Low, None; S. Albani, None.
Background/Purpose: Systemic lupus erythematosus (SLE) is a complex, systemic autoimmune disease that interferes with the balance between regulation and immunity, resulting in immune system dysfunction. Disease course is unpredictable due to alternating remissions and flares, impeding reliable patient assessments. Thus, mechanistic insights are required for better assessment. Current studies are mainly descriptive, and SLE is best interrogated with a multi-parametric, holistic approach such as mass cytometry (CyTOF).
The objectives of this study are as follows: (1) To characterise immune signatures of SLE patients and in the process: Study the roles of B and T cells in SLE to gain a holistic understanding of the adaptive immune response and (2) To compare immunological profiles of SLE patients with age-matched healthy controls.
Methods: Peripheral blood mononuclear cells (PBMCs) from 26 SLE subjects (41 samples, median age 39.5years) and 27 age-matched subjects were tested with CyTOF. Data was analysed and visualised using our laboratory-designed machine learning tool, the Extended Polydimensional Immunome Characterization (EPIC) platform. Normalization and FlowSOM (Flow cytometry analysis by Self- Organising Maps) clustering were performed with functionally and phenotypically important immune markers. Mann-Whitney U test identified significantly different cluster frequencies.
Results: Multiple significant differences were identified (p < 0.05). An activated CD8+CXCR3+CLA+ T -cell subset (CD8+CD45RA+CD38+CLA+CXCR3+) was enriched in SLE (median: 0.19%,interquartile range: 0.13-0.35% of CD45+ PBMCs) versus healthy (0.08%,0.05-0.09%; p< 0.0001). CXCR3 and cutaneous lymphocyte-associated antigen (CLA) are involved in skin-homing, with CLA function enhanced in inflammatory dermatoses. This suggests the importance of skin involvement in SLE immunopathogenesis even if skin manifestations are absent. Further analysis of CD3+ cells revealed unchanged natural T-regulatory cells (TREGS) in healthy and disease while a few TREG-like populations (FoxP3+CD25-) are significantly increased in disease, suggesting a deranged immunoregulatory response driven by TREGS. Thirdly, an activated CD8+BAFF+ T-cell subset (CD8+CD45RA+iCOS+ BAFF+) (r=0.715, p< 0.0001) is enriched in SLE (0.975%,0.433-1.53%) versus healthy (0.28%,0.1-0.49%; p< 0.0001). B-cell activating factor (BAFF) supports autoreactive B cell survival in autoimmune disease; an anti-BAFF drug (belimumab) is FDA- approved for SLE. Despite increasing use of belimumab, not all patients respond equally, so BAFF inhibition alone may not adequately alter disease activity. Studying these cell subset interactions in further detail to identify pathological pathways may facilitate improvements in current SLE theragnostics.
Conclusion: With a multi-parametric unbiased approach comparing SLE subjects to healthy controls, we identified immune subsets of immunopathogenic importance for further studies.
 Figure 1: Clinical samples are used for experimentation and subsequent analysis.
Figure 1: Clinical samples are used for experimentation and subsequent analysis.  Figure 2: A graphic representation of the EPIC Immunomics Platform, which provides a healthy dataset and analysis pipeline to identify derangements in disease.
Figure 2: A graphic representation of the EPIC Immunomics Platform, which provides a healthy dataset and analysis pipeline to identify derangements in disease.  Figure 3: There are derangements in multiple subsets in SLE. Notably, many differences between SLE and healthy were observed in the CD3+ compartment (circled in red).
Figure 3: There are derangements in multiple subsets in SLE. Notably, many differences between SLE and healthy were observed in the CD3+ compartment (circled in red). Disclosures: K. Nay Yaung, None; J. Yeo, None; H. Law, None; M. Wasser, None; T. Arkachaisri, None; J. Thumboo, None; A. Low, None; S. Albani, None.

