Back
Abstract Session
Periodic fever syndromes, autoinflammatory diseases, Still’s disease and MAS/HLH
Session: Abstracts: Pediatric Rheumatology – Basic Science (0566–0569)
0566: SARS-CoV-2-Specific T Cell Responses in Multisystem Inflammatory Syndrome in Children
Sunday, November 13, 2022
8:00 AM – 8:10 AM Eastern Time
Location: Ballroom AB
- KL
Ki Pui Lam, PhD
Division of Immunology, Boston Childrens Hospital, Harvard Medical School
Boston, MA, United States
Presenting Author(s)
Ki Pui Lam1, Marcos H. Chinas2, Amélie M. Julé2, Maria Taylor3, Mary Beth F. Son1, Janet Chou2, Jane W. Newburger4, Adrienne G. Randolph5, Maria Gutierrez-Arcelus2 and lauren henderson1, 1Division of Immunology, Boston Children's Hospital, Boston, MA, 2Division of Immunology, Boston Children's Hospital, Harvard Medical School, Boston, MA, 3Division of Immunology, Boston Children's Hospital, Brighton, MA, 4Department of Cardiology, Boston Children's Hospital, Harvard Medical School, Boston, MA, 5Division of Critical Care Medicine, Boston Children's Hospital, Harvard Medical School, Boston, MA
Background/Purpose: Multisystem inflammatory syndrome in children (MIS-C) develops about a month after SARS-CoV-2 infection and this delayed presentation suggests a role for the adaptive immune system. While the majority of children with MIS-C generate antibodies that neutralize SARS-CoV-2, less is known about antigen-specific T cells. We sought to characterize T cell responses in MIS-C compared to COVID-19 and pediatric hyperinflammatory syndromes.
Methods: Children with MIS-C (CDC or WHO case definitions) and COVID-19 were recruited from our center (5/2020-4/2021) and provided blood samples. Severe MIS-C was defined as ICU admission or development of coronary artery aneurysms. Children with systemic juvenile idiopathic arthritis (ILAR criteria), macrophage activation syndrome (MAS) (2016 MAS in sJIA Classification Criteria), Kawasaki disease (American Heart Association criteria), and fever unrelated to SARS-CoV-2 served as comparison subjects. To characterize the T cell repertoire, genomic DNA was used to sequence the T cell receptor (TCR) beta chain (Adaptive Biotechnologies). HLA-typing was performed by the Red Cross. The ImmuneCODE database was sourced to provide T cell repertoire data from adults with COVID-19 and TCR sequences that recognize SARS-CoV-2. Antigen specific T cells were identified by stimulating T cells with SARS-CoV-2 peptides and measuring CD40 ligand (CD40L) expression by flow cytometry.
Results: Clinical characteristics of the patients are in Table 1. TRBV11-2 gene usage was increased in MIS-C patients who tested positive for SARS-CoV-2 (MIS-Cpos, n=19) compared to all other pediatric cohorts (p< 0.0001) and adults with COVID-19 in the ImmuneCODE database (Fig1). A small number of adults (4/159) in the COVID-19 recovered group had increased Vβ11-2+ T cells. Children diagnosed with MIS-C by their providers but were negative for SARS-CoV-2 by PCR and serology (MIS-Cneg, n=5) did not display Vβ skewing. Disease severity, prior immunomodulatory treatment, age, time to sample collection, and HLA typing were not significantly associated with expansion of Vβ11-2+T cells in MIS-Cpos patients (Fig1). In T cells expressing TRBV11-2, Jβ gene pairing was polyclonal. There was a trend towards expanded SARS-CoV-2-specific TCRs in MIS-Cpos patients that was statistical significant when compared to febrile controls (p=0.03)(Fig2). TRBV11-2 gene usage was not enriched in SARS-CoV-2-specific TCRs of MIS-Cpos patients when compared to pediatric COVID-19. The viral antigens recognized by SARS-CoV-2-specific TCRs were similar in children with MIS-C and COVID-19 (Fig2). CD40L+ T cells were identified in children with MIS-Cpos after stimulation with SARS-CoV-2 peptides at frequencies comparable to children with convalescent COVID-19 (Fig 2). There was no difference in the frequency of CD40L+ T cells in MIS-Cpos patients based on disease severity.
Conclusion: Expansion of Vβ11-2+ T cells was a specific biomarker of MIS-C patients with laboratory confirmed SARS-CoV-2 infections and was not found in COVID-19 or pediatric hyperinflammatory syndromes. Children with MIS-C had robust antigen-specific T cell responses to SARS-CoV-2 that were comparable to pediatric patients with COVID-19.
.jpg)
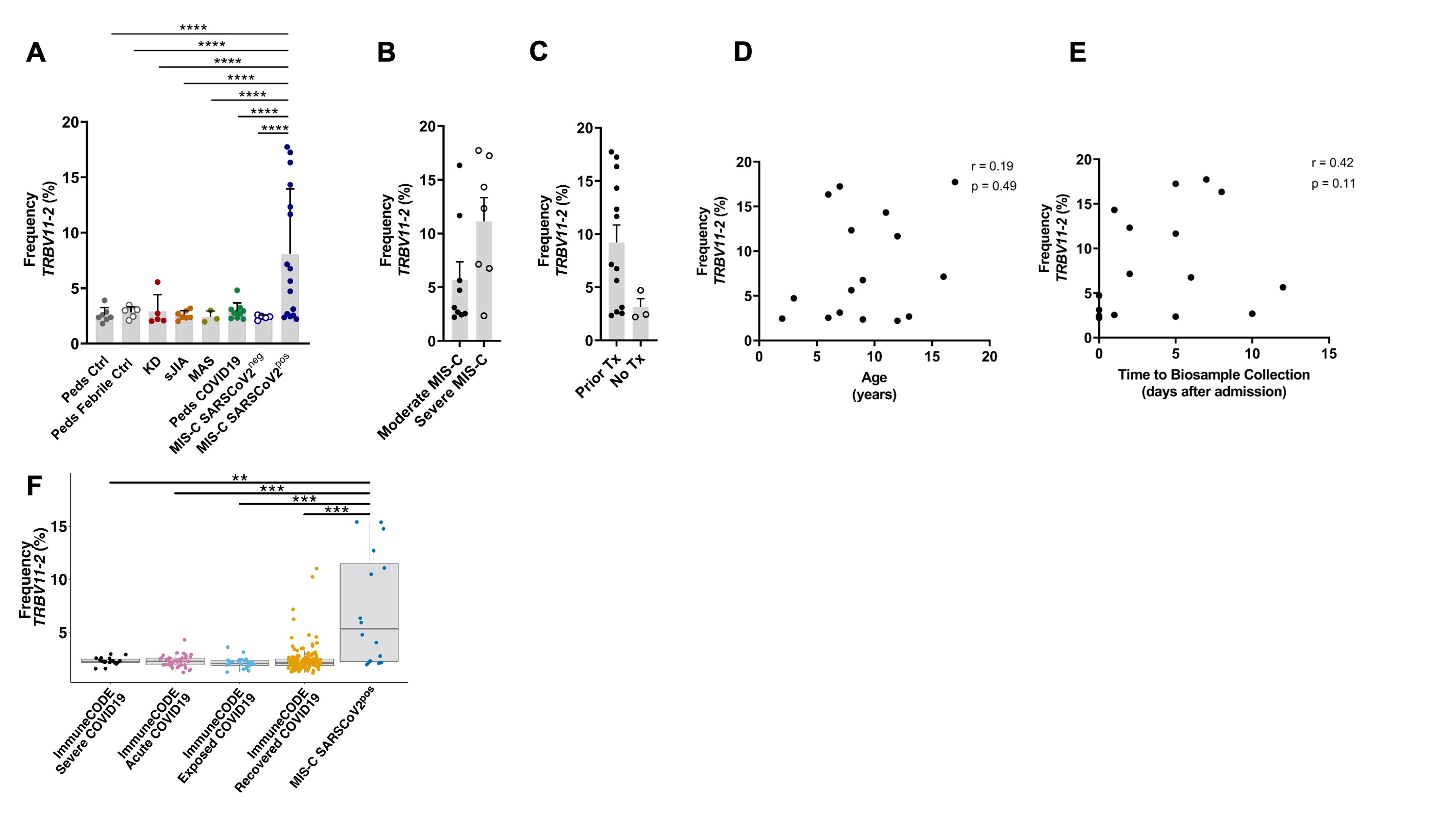 Figure 1. Increased Usage of TRBV11-2 in T cells from patients with MIS-C. A) The frequency of T cell clonotypes expressing TRBV11-2 in each study group. Statistical analysis: two-way ANOVA comparing study group and Vβ gene with the Dunnett’s multiple comparisons test. The frequency of T cell clonotypes expressing TRBV11-2 in patients with B) moderate vs. severeª MIS-C SARSCoV2pos and C) patients with MIS-C SARSCoV2pos who received prior immunomodulatory treatment vs. those who were treatment naïve. The relationship between the frequency of T cell clonotypes expressing TRBV11-2 and D) the age and E) time from hospital admission to biosample collection in patients with MIS-C SARSCoV2pos. Statistical analysis: Spearman’s non-parametric rank correlation. F) The frequency of T cell clonotypes expressing TRBV11-2 in MIS-C SARSCoV2pos and adults in the ImmuneCODE database. Statistical analysis: Wilcoxon test. Summary data on bar graphs is mean ± standard error in A-E and median with IQR in F. P value, **** ≤ 0.0001, ***=0.001, **=0.01, *=0.05.
Figure 1. Increased Usage of TRBV11-2 in T cells from patients with MIS-C. A) The frequency of T cell clonotypes expressing TRBV11-2 in each study group. Statistical analysis: two-way ANOVA comparing study group and Vβ gene with the Dunnett’s multiple comparisons test. The frequency of T cell clonotypes expressing TRBV11-2 in patients with B) moderate vs. severeª MIS-C SARSCoV2pos and C) patients with MIS-C SARSCoV2pos who received prior immunomodulatory treatment vs. those who were treatment naïve. The relationship between the frequency of T cell clonotypes expressing TRBV11-2 and D) the age and E) time from hospital admission to biosample collection in patients with MIS-C SARSCoV2pos. Statistical analysis: Spearman’s non-parametric rank correlation. F) The frequency of T cell clonotypes expressing TRBV11-2 in MIS-C SARSCoV2pos and adults in the ImmuneCODE database. Statistical analysis: Wilcoxon test. Summary data on bar graphs is mean ± standard error in A-E and median with IQR in F. P value, **** ≤ 0.0001, ***=0.001, **=0.01, *=0.05.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; KD, Kawasaki disease; sJIA, systemic juvenile idiopathic arthritis; MAS, macrophage activation syndrome; Tx, treatment.
ª Severe MIS-C was defined by admission to the intensive care unit or development of coronary artery aneurysms (CAAs) (z-score ≥ 2.5).
.jpg) Figure 2. SARS-CoV-2 Specific T Cell Responses in MIS-C. SARS-CoV-2 specific TCR sequences were obtained from the ImmuneCODE database. Antigen-specific T cells were identified by the expression of activation markers following exposure to SARS-CoV-2 peptides. These T cells were then isolated for TCR sequencing. A) The median frequency with IQR of the top 10 most abundant SARS-CoV-2-specific TCRs. B-C) Viral antigens recognized by SARS-CoV-2-specific TCRs in B) MIS-C SARSCOV2pos and C) pediatric COVID-19. D-E) Peripheral blood mononuclear cells were incubated with either water (negative control) or peptide pools covering the S1, N, and M viral proteins. SARS-CoV-2 -specific T cells were identified by their expression of CD154 (CD40 ligand). D) Flow cytometry dot plots depicting CD40L expression on T cells from a healthy control, child with convalescent COVID-19ª, and child with MIS-C SARSCOV2pos. E) Frequency of CD4+CD40L+ or CD8+CD40L+ T cells in the given study group. Summary data on bar graphs is mean ± standard error. P value * ≤ 0.05, ** ≤ 0.01.
Figure 2. SARS-CoV-2 Specific T Cell Responses in MIS-C. SARS-CoV-2 specific TCR sequences were obtained from the ImmuneCODE database. Antigen-specific T cells were identified by the expression of activation markers following exposure to SARS-CoV-2 peptides. These T cells were then isolated for TCR sequencing. A) The median frequency with IQR of the top 10 most abundant SARS-CoV-2-specific TCRs. B-C) Viral antigens recognized by SARS-CoV-2-specific TCRs in B) MIS-C SARSCOV2pos and C) pediatric COVID-19. D-E) Peripheral blood mononuclear cells were incubated with either water (negative control) or peptide pools covering the S1, N, and M viral proteins. SARS-CoV-2 -specific T cells were identified by their expression of CD154 (CD40 ligand). D) Flow cytometry dot plots depicting CD40L expression on T cells from a healthy control, child with convalescent COVID-19ª, and child with MIS-C SARSCOV2pos. E) Frequency of CD4+CD40L+ or CD8+CD40L+ T cells in the given study group. Summary data on bar graphs is mean ± standard error. P value * ≤ 0.05, ** ≤ 0.01.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; TCR, T cell receptor.
ª Blood samples obtained greater than 2 weeks after a positive PCR for SARS-CoV-2 were classified as convalescent COVID-19.
Disclosures: K. Lam, None; M. Chinas, None; A. Julé, None; M. Taylor, None; M. Son, None; J. Chou, None; J. Newburger, None; A. Randolph, None; M. Gutierrez-Arcelus, None; l. henderson, Sobi, Pfizer, Adaptive Biotechnologies, BMS.
Background/Purpose: Multisystem inflammatory syndrome in children (MIS-C) develops about a month after SARS-CoV-2 infection and this delayed presentation suggests a role for the adaptive immune system. While the majority of children with MIS-C generate antibodies that neutralize SARS-CoV-2, less is known about antigen-specific T cells. We sought to characterize T cell responses in MIS-C compared to COVID-19 and pediatric hyperinflammatory syndromes.
Methods: Children with MIS-C (CDC or WHO case definitions) and COVID-19 were recruited from our center (5/2020-4/2021) and provided blood samples. Severe MIS-C was defined as ICU admission or development of coronary artery aneurysms. Children with systemic juvenile idiopathic arthritis (ILAR criteria), macrophage activation syndrome (MAS) (2016 MAS in sJIA Classification Criteria), Kawasaki disease (American Heart Association criteria), and fever unrelated to SARS-CoV-2 served as comparison subjects. To characterize the T cell repertoire, genomic DNA was used to sequence the T cell receptor (TCR) beta chain (Adaptive Biotechnologies). HLA-typing was performed by the Red Cross. The ImmuneCODE database was sourced to provide T cell repertoire data from adults with COVID-19 and TCR sequences that recognize SARS-CoV-2. Antigen specific T cells were identified by stimulating T cells with SARS-CoV-2 peptides and measuring CD40 ligand (CD40L) expression by flow cytometry.
Results: Clinical characteristics of the patients are in Table 1. TRBV11-2 gene usage was increased in MIS-C patients who tested positive for SARS-CoV-2 (MIS-Cpos, n=19) compared to all other pediatric cohorts (p< 0.0001) and adults with COVID-19 in the ImmuneCODE database (Fig1). A small number of adults (4/159) in the COVID-19 recovered group had increased Vβ11-2+ T cells. Children diagnosed with MIS-C by their providers but were negative for SARS-CoV-2 by PCR and serology (MIS-Cneg, n=5) did not display Vβ skewing. Disease severity, prior immunomodulatory treatment, age, time to sample collection, and HLA typing were not significantly associated with expansion of Vβ11-2+T cells in MIS-Cpos patients (Fig1). In T cells expressing TRBV11-2, Jβ gene pairing was polyclonal. There was a trend towards expanded SARS-CoV-2-specific TCRs in MIS-Cpos patients that was statistical significant when compared to febrile controls (p=0.03)(Fig2). TRBV11-2 gene usage was not enriched in SARS-CoV-2-specific TCRs of MIS-Cpos patients when compared to pediatric COVID-19. The viral antigens recognized by SARS-CoV-2-specific TCRs were similar in children with MIS-C and COVID-19 (Fig2). CD40L+ T cells were identified in children with MIS-Cpos after stimulation with SARS-CoV-2 peptides at frequencies comparable to children with convalescent COVID-19 (Fig 2). There was no difference in the frequency of CD40L+ T cells in MIS-Cpos patients based on disease severity.
Conclusion: Expansion of Vβ11-2+ T cells was a specific biomarker of MIS-C patients with laboratory confirmed SARS-CoV-2 infections and was not found in COVID-19 or pediatric hyperinflammatory syndromes. Children with MIS-C had robust antigen-specific T cell responses to SARS-CoV-2 that were comparable to pediatric patients with COVID-19.
.jpg)
 Figure 1. Increased Usage of TRBV11-2 in T cells from patients with MIS-C. A) The frequency of T cell clonotypes expressing TRBV11-2 in each study group. Statistical analysis: two-way ANOVA comparing study group and Vβ gene with the Dunnett’s multiple comparisons test. The frequency of T cell clonotypes expressing TRBV11-2 in patients with B) moderate vs. severeª MIS-C SARSCoV2pos and C) patients with MIS-C SARSCoV2pos who received prior immunomodulatory treatment vs. those who were treatment naïve. The relationship between the frequency of T cell clonotypes expressing TRBV11-2 and D) the age and E) time from hospital admission to biosample collection in patients with MIS-C SARSCoV2pos. Statistical analysis: Spearman’s non-parametric rank correlation. F) The frequency of T cell clonotypes expressing TRBV11-2 in MIS-C SARSCoV2pos and adults in the ImmuneCODE database. Statistical analysis: Wilcoxon test. Summary data on bar graphs is mean ± standard error in A-E and median with IQR in F. P value, **** ≤ 0.0001, ***=0.001, **=0.01, *=0.05.
Figure 1. Increased Usage of TRBV11-2 in T cells from patients with MIS-C. A) The frequency of T cell clonotypes expressing TRBV11-2 in each study group. Statistical analysis: two-way ANOVA comparing study group and Vβ gene with the Dunnett’s multiple comparisons test. The frequency of T cell clonotypes expressing TRBV11-2 in patients with B) moderate vs. severeª MIS-C SARSCoV2pos and C) patients with MIS-C SARSCoV2pos who received prior immunomodulatory treatment vs. those who were treatment naïve. The relationship between the frequency of T cell clonotypes expressing TRBV11-2 and D) the age and E) time from hospital admission to biosample collection in patients with MIS-C SARSCoV2pos. Statistical analysis: Spearman’s non-parametric rank correlation. F) The frequency of T cell clonotypes expressing TRBV11-2 in MIS-C SARSCoV2pos and adults in the ImmuneCODE database. Statistical analysis: Wilcoxon test. Summary data on bar graphs is mean ± standard error in A-E and median with IQR in F. P value, **** ≤ 0.0001, ***=0.001, **=0.01, *=0.05. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; KD, Kawasaki disease; sJIA, systemic juvenile idiopathic arthritis; MAS, macrophage activation syndrome; Tx, treatment.
ª Severe MIS-C was defined by admission to the intensive care unit or development of coronary artery aneurysms (CAAs) (z-score ≥ 2.5).
.jpg) Figure 2. SARS-CoV-2 Specific T Cell Responses in MIS-C. SARS-CoV-2 specific TCR sequences were obtained from the ImmuneCODE database. Antigen-specific T cells were identified by the expression of activation markers following exposure to SARS-CoV-2 peptides. These T cells were then isolated for TCR sequencing. A) The median frequency with IQR of the top 10 most abundant SARS-CoV-2-specific TCRs. B-C) Viral antigens recognized by SARS-CoV-2-specific TCRs in B) MIS-C SARSCOV2pos and C) pediatric COVID-19. D-E) Peripheral blood mononuclear cells were incubated with either water (negative control) or peptide pools covering the S1, N, and M viral proteins. SARS-CoV-2 -specific T cells were identified by their expression of CD154 (CD40 ligand). D) Flow cytometry dot plots depicting CD40L expression on T cells from a healthy control, child with convalescent COVID-19ª, and child with MIS-C SARSCOV2pos. E) Frequency of CD4+CD40L+ or CD8+CD40L+ T cells in the given study group. Summary data on bar graphs is mean ± standard error. P value * ≤ 0.05, ** ≤ 0.01.
Figure 2. SARS-CoV-2 Specific T Cell Responses in MIS-C. SARS-CoV-2 specific TCR sequences were obtained from the ImmuneCODE database. Antigen-specific T cells were identified by the expression of activation markers following exposure to SARS-CoV-2 peptides. These T cells were then isolated for TCR sequencing. A) The median frequency with IQR of the top 10 most abundant SARS-CoV-2-specific TCRs. B-C) Viral antigens recognized by SARS-CoV-2-specific TCRs in B) MIS-C SARSCOV2pos and C) pediatric COVID-19. D-E) Peripheral blood mononuclear cells were incubated with either water (negative control) or peptide pools covering the S1, N, and M viral proteins. SARS-CoV-2 -specific T cells were identified by their expression of CD154 (CD40 ligand). D) Flow cytometry dot plots depicting CD40L expression on T cells from a healthy control, child with convalescent COVID-19ª, and child with MIS-C SARSCOV2pos. E) Frequency of CD4+CD40L+ or CD8+CD40L+ T cells in the given study group. Summary data on bar graphs is mean ± standard error. P value * ≤ 0.05, ** ≤ 0.01.SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MIS-C, multisystem inflammatory syndrome in children; peds, pediatric; ctrl, control; TCR, T cell receptor.
ª Blood samples obtained greater than 2 weeks after a positive PCR for SARS-CoV-2 were classified as convalescent COVID-19.
Disclosures: K. Lam, None; M. Chinas, None; A. Julé, None; M. Taylor, None; M. Son, None; J. Chou, None; J. Newburger, None; A. Randolph, None; M. Gutierrez-Arcelus, None; l. henderson, Sobi, Pfizer, Adaptive Biotechnologies, BMS.

