Back
Poster Session B
Systemic lupus erythematosus (SLE)
Session: (0629–0670) SLE – Etiology and Pathogenesis Poster
0667: Correlation Matrices Visualize Differential Degree of Cell and Pathway Heterogeneity in Skin of Cutaneous Lupus Erythematosus Treatment Subgroups
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- FC
Felix Chin, BS
University of Pennsylvania Perelman School of Medicine
Philadelphia, PA, United States
Abstract Poster Presenter(s)
Felix Chin1, Thomas Vazquez2, Josh Dan3, DeAnna Diaz4, Grant Sprow5, Jay Patel6, Nilesh Kodali7, Rui Feng8 and Victoria Werth9, 1University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, 2FIU Wertheim College of Medicine, Virginia Beach, VA, 3Philadelphia VAMC, Philadelphia, PA, USA and Department of Dermatology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, 4Philadelphia College of Medicine, Philadelphia, PA, 5Albert Einstein College of Medicine, Philadelphia, PA, 6Philadelphia VAMC, Philadelphia, PA, USA and Department of Dermatology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PN, 7New Jersey Medical School, Coppell, TX, 8University of Pennsylvania, Philadelphia, 9University of Pennsylvania and Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA
Background/Purpose: First-line treatment for cutaneous lupus erythematosus involves the use of antimalarials. Treatment response is highly variable with some patients responding well to hydroxychloroquine (HCQ), some necessitating the addition of quinacrine (QC) and others refractory to treatment from both (NR). Here we compared the immune infiltrate composition of CLE skin between 3 different treatment response groups with imaging mass cytometry. We then used correlation matrices to highlight the differences in pathway heterogeneity between each of the groups. We also employed unsupervised learning to visualize the distinct cell-dominant subtypes in each of the groups.
Methods: Infiltrate composition of CLE skin biopsies were characterized using imaging mass cytometry, where we stained biopsies (12 HCQ, 11 QC, 20 NR) with two separate panels of 37-metal conjugated antibodies, which enabled the measurement of different cell types, cytokines and pathway proteins within the tissue sample. Correlation matrices of cytokines/pathways with superimposed hierarchical clustering were constructed for each of the three treatment groups to visualize correlation strength for all markers simultaneously. Cell measurements were scaled within each cell type and then subjected to K-means clustering. Results of cell clustering were visualized using horizontally scaled heat maps, which demonstrated cellular predominance within each patient's biopsy.
Results: Correlation matrices revealed a single large correlation cluster in HCQ, two medium clusters in QC and many clusters in NR. This indicates relatively homogenous cytokine/pathway activity in HCQ, two separate mechanisms in QC and significant heterogeneity in NR. Cell clustering heatmaps demonstrated heavy CD4/CD8 dominance in HCQ, two noteworthy QC subgroups, one being dominated by CD8 cells (61% vs. 7%, p=0.1) and the other by cDC (62% vs. 15%, p=0.1), and three noteworthy NR subgroups being CD8-dominant (48% vs. 9%, p< 0.00001), cDC-dominant (38% vs. 10%, p< 0.001) and CD14+CD16+ dominant (45% vs. 4%, p< 0.00001). Furthermore, CD4/CD8 dominance in HCQ appeared to be positively associated with general increased levels of pathways.
Conclusion:
HCQ responsive patients may share a common origin for pathway activity whereas QC patients potentially possess 2 separate mechanisms responsive to quinacrine therapy. NR patients showed substantial heterogeneity, which implies multiple possible origins of pathway activity thus supporting the need for personalized therapy. In terms of cells, tandem CD4/CD8 presence could potentially be used as a predictor for hydroxychloroquine-responsiveness. In QC and NR subgroups, most patients displayed single cell type predominance, most notably cDC and CD8, which suggests potential utility for personalized medicine in targeting single cell types.
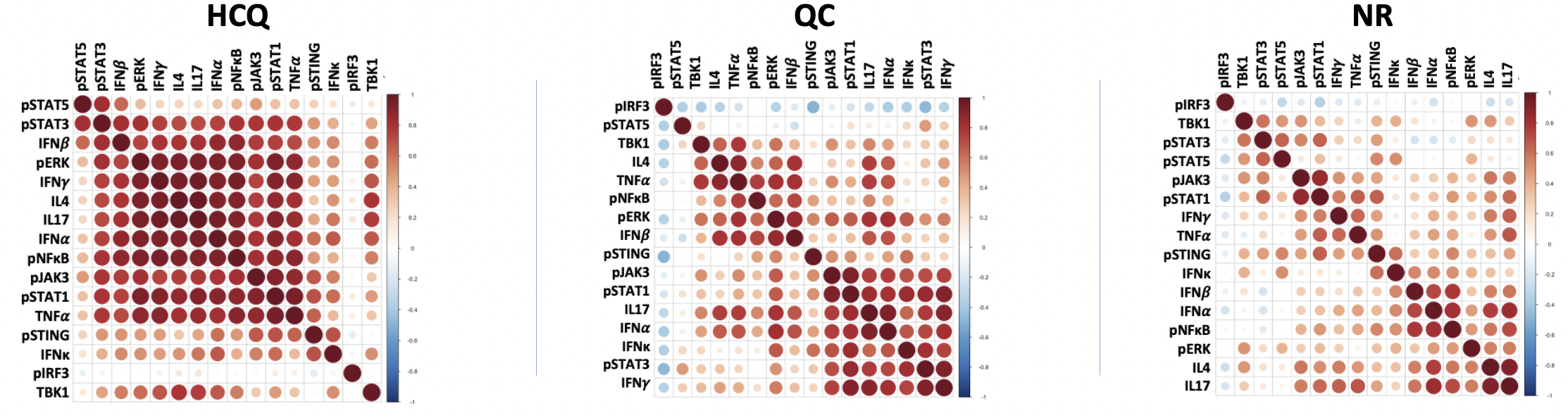 Fig 1. Size of each dot is a reflection of correlation strength (red = positive). Correlations were grouped using hierarchical clustering. HCQ shows single large cluster indicating pathways coexist together homogeneously. QC shows two main clusters suggesting two potential mechanisms responsive to quinacrine. NR shows no clusters indicating significant heterogeneity.
Fig 1. Size of each dot is a reflection of correlation strength (red = positive). Correlations were grouped using hierarchical clustering. HCQ shows single large cluster indicating pathways coexist together homogeneously. QC shows two main clusters suggesting two potential mechanisms responsive to quinacrine. NR shows no clusters indicating significant heterogeneity.
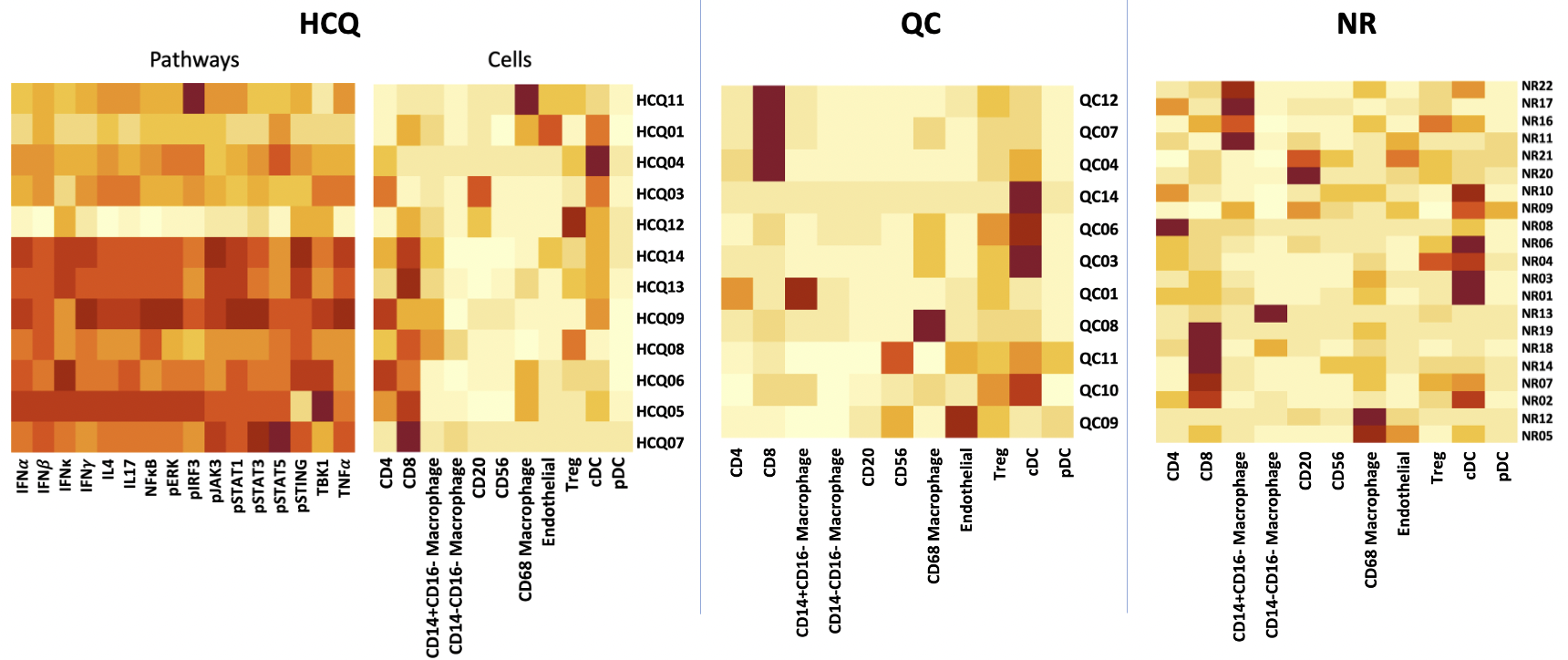 Fig 1. K-means clustered heatmaps illustrating the different cell clusters that exist within the three treatment response groups. Each row represents a single patient. Greater CD4/CD8 activity appeared to correlate with general pathway activity in the HCQ group. QC and NR clusters contain similarities in terms of single-cell dominance, both showing CD8 and cDC-predominant subgroups..
Fig 1. K-means clustered heatmaps illustrating the different cell clusters that exist within the three treatment response groups. Each row represents a single patient. Greater CD4/CD8 activity appeared to correlate with general pathway activity in the HCQ group. QC and NR clusters contain similarities in terms of single-cell dominance, both showing CD8 and cDC-predominant subgroups..
Disclosures: F. Chin, None; T. Vazquez, None; J. Dan, None; D. Diaz, None; G. Sprow, None; J. Patel, None; N. Kodali, None; R. Feng, None; V. Werth, AbbVie/Abbott, Amgen, AstraZeneca, Bristol Myers Squibb (BMS), Celgene, Eli Lilly, Genentech, GlaxoSmithKline (GSK), Janssen, Merck/MSD, Gilead, Novartis, Pfizer, Rome Pharmaceuticals, Horizon Therapeutics, Regeneron, argenx, CSL Behring, AnaptysBio, Biogen, Corbus, EMD Serono, Galderma, Nektar, Octapharma, Principia, Resolve, Sanofi, Syntimmune, Viela.
Background/Purpose: First-line treatment for cutaneous lupus erythematosus involves the use of antimalarials. Treatment response is highly variable with some patients responding well to hydroxychloroquine (HCQ), some necessitating the addition of quinacrine (QC) and others refractory to treatment from both (NR). Here we compared the immune infiltrate composition of CLE skin between 3 different treatment response groups with imaging mass cytometry. We then used correlation matrices to highlight the differences in pathway heterogeneity between each of the groups. We also employed unsupervised learning to visualize the distinct cell-dominant subtypes in each of the groups.
Methods: Infiltrate composition of CLE skin biopsies were characterized using imaging mass cytometry, where we stained biopsies (12 HCQ, 11 QC, 20 NR) with two separate panels of 37-metal conjugated antibodies, which enabled the measurement of different cell types, cytokines and pathway proteins within the tissue sample. Correlation matrices of cytokines/pathways with superimposed hierarchical clustering were constructed for each of the three treatment groups to visualize correlation strength for all markers simultaneously. Cell measurements were scaled within each cell type and then subjected to K-means clustering. Results of cell clustering were visualized using horizontally scaled heat maps, which demonstrated cellular predominance within each patient's biopsy.
Results: Correlation matrices revealed a single large correlation cluster in HCQ, two medium clusters in QC and many clusters in NR. This indicates relatively homogenous cytokine/pathway activity in HCQ, two separate mechanisms in QC and significant heterogeneity in NR. Cell clustering heatmaps demonstrated heavy CD4/CD8 dominance in HCQ, two noteworthy QC subgroups, one being dominated by CD8 cells (61% vs. 7%, p=0.1) and the other by cDC (62% vs. 15%, p=0.1), and three noteworthy NR subgroups being CD8-dominant (48% vs. 9%, p< 0.00001), cDC-dominant (38% vs. 10%, p< 0.001) and CD14+CD16+ dominant (45% vs. 4%, p< 0.00001). Furthermore, CD4/CD8 dominance in HCQ appeared to be positively associated with general increased levels of pathways.
Conclusion:
HCQ responsive patients may share a common origin for pathway activity whereas QC patients potentially possess 2 separate mechanisms responsive to quinacrine therapy. NR patients showed substantial heterogeneity, which implies multiple possible origins of pathway activity thus supporting the need for personalized therapy. In terms of cells, tandem CD4/CD8 presence could potentially be used as a predictor for hydroxychloroquine-responsiveness. In QC and NR subgroups, most patients displayed single cell type predominance, most notably cDC and CD8, which suggests potential utility for personalized medicine in targeting single cell types.
 Fig 1. Size of each dot is a reflection of correlation strength (red = positive). Correlations were grouped using hierarchical clustering. HCQ shows single large cluster indicating pathways coexist together homogeneously. QC shows two main clusters suggesting two potential mechanisms responsive to quinacrine. NR shows no clusters indicating significant heterogeneity.
Fig 1. Size of each dot is a reflection of correlation strength (red = positive). Correlations were grouped using hierarchical clustering. HCQ shows single large cluster indicating pathways coexist together homogeneously. QC shows two main clusters suggesting two potential mechanisms responsive to quinacrine. NR shows no clusters indicating significant heterogeneity. Fig 1. K-means clustered heatmaps illustrating the different cell clusters that exist within the three treatment response groups. Each row represents a single patient. Greater CD4/CD8 activity appeared to correlate with general pathway activity in the HCQ group. QC and NR clusters contain similarities in terms of single-cell dominance, both showing CD8 and cDC-predominant subgroups..
Fig 1. K-means clustered heatmaps illustrating the different cell clusters that exist within the three treatment response groups. Each row represents a single patient. Greater CD4/CD8 activity appeared to correlate with general pathway activity in the HCQ group. QC and NR clusters contain similarities in terms of single-cell dominance, both showing CD8 and cDC-predominant subgroups.. Disclosures: F. Chin, None; T. Vazquez, None; J. Dan, None; D. Diaz, None; G. Sprow, None; J. Patel, None; N. Kodali, None; R. Feng, None; V. Werth, AbbVie/Abbott, Amgen, AstraZeneca, Bristol Myers Squibb (BMS), Celgene, Eli Lilly, Genentech, GlaxoSmithKline (GSK), Janssen, Merck/MSD, Gilead, Novartis, Pfizer, Rome Pharmaceuticals, Horizon Therapeutics, Regeneron, argenx, CSL Behring, AnaptysBio, Biogen, Corbus, EMD Serono, Galderma, Nektar, Octapharma, Principia, Resolve, Sanofi, Syntimmune, Viela.

