Back
Poster Session B
Rheumatoid arthritis (RA)
Session: (0913–0938) RA – Treatment Poster II
0933: Exploratory Study of the Usefulness of TNFAIP3 Genetic Variants in Predicting Response to Methotrexate in Early Arthritis Patients
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- AT
Ana Triguero Martínez, BSc
Hospital Universitario La Princesa
Madrid, Spain
Abstract Poster Presenter(s)
Ana Triguero1, JUAN BALDIVIESO ACHA2, Emilia Roy Vallejo3, Patricia Quiroga-Colina4, Irene Maria Llorente Cubas2, Rosario Garcia-Vicuña5, amaya Puig kröger6 and Isidoro Gonzalez2, 1Hospital Universitario La Princesa, Madrid, Spain, 2Hospital Universitario de La Princesa, Madrid, Spain, 3Internal Medicine Department, Hospital Universitario La Princesa, Instituto de Investigación Sanitaria La Princesa (IIS-IP), Madrid, Spain, 4Division of Rheumatology, Hospital Universitario de La Princesa, Madrid, Spain, 5Hospital Universitario de La Princesa, IIS-Princesa, Madrid, Spain, 6Laboratory of Immunometabolism, Hospital General Universitario Gregorio Marañón, Madrid, Spain
Background/Purpose: We recently described that methotrexate (MTX) could exert its immunomodulatory effect through the induction of A20 expression , a protein that negatively regulates NFkB signalling induced by TLR ligands or TNF stimulation. On the other hand, different single nucleotide polymorphisms (SNPs) in TNFAIP3, the gene that codes the A20 protein, have been associated with an increased risk to develop rheumatoid arthritis (RA). It is likely that these variants could be associated with lower expression and activity of A20.This study aims to determine whether those SNPs in TNFAIP3, associated with increased risk of RA, are useful for predicting MTX response.
Methods: We analysed data from PEARL study (Princesa Early Arthritis Register Longitudinal study; CEIC approval registry no 518; March 2011) in which sociodemographic, clinical, analytical, therapeutic variables and biological samples are collect by protocol in 5 structured visits.
We assessed clinical response (no, moderate and good) at six months in those patients who started MTX at the baseline and maintained this drug in monotherapy, by using the EULAR and HUPI response criteria. The following SNPs in TNFAIP3, rs600144, rs11970361, rs582757, rs11970411, and rs17780429, were genotyped using TaqMan probes (ThermoFisher Scientific; part number:C_7701163_10, C_238210_20, C_8300291_10, C_32241642_10, C_34723620_10, respectively).
To determine whether there was an association between the response to MTX and the SNPs, an order logistic regression model was performed using the ologit command of Stata 14.1. The model was adjusted for age, sex, MTX dose (median 15 mg; interquartile range 12.5 - 20) and disease activity at baseline.
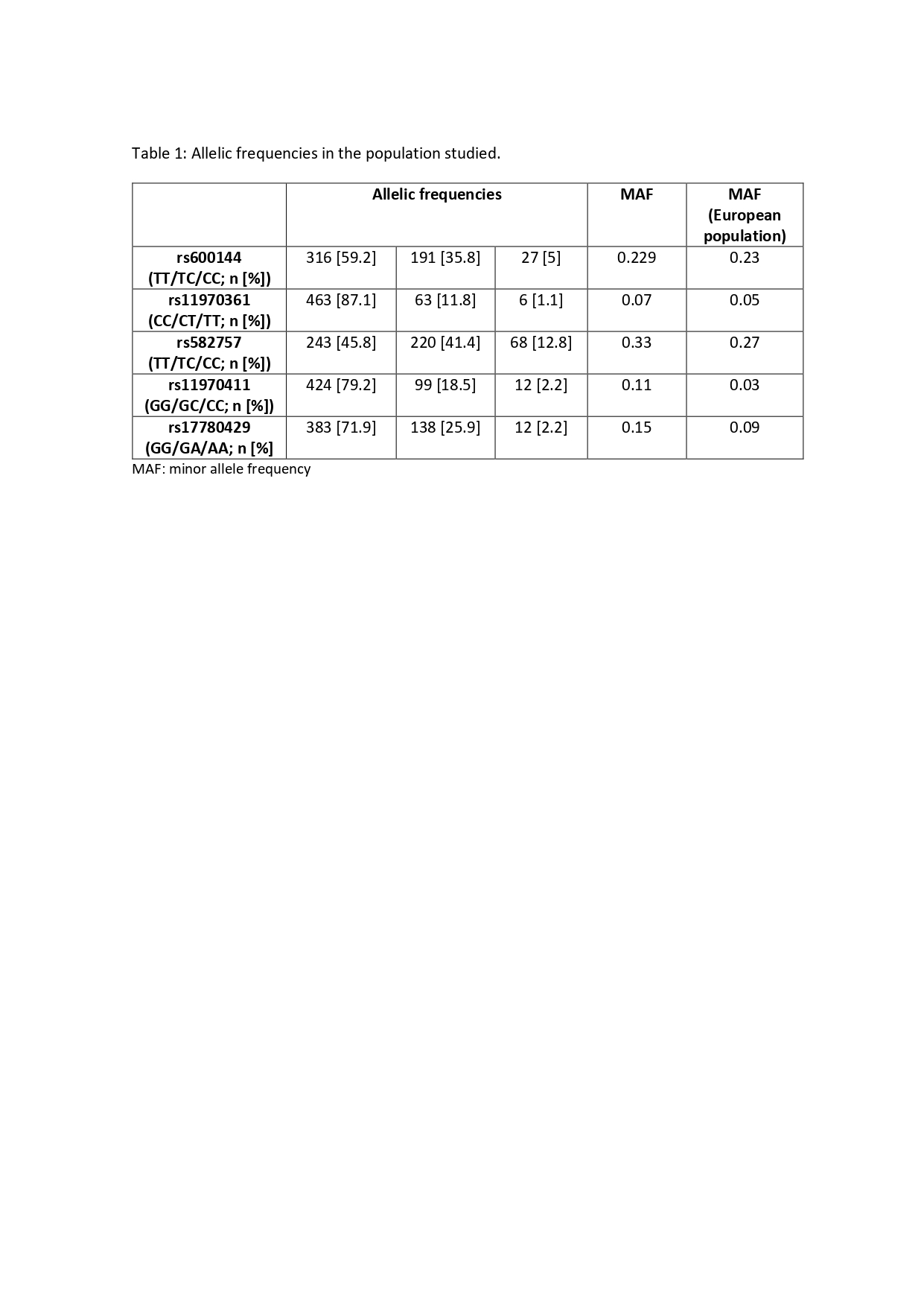
Results: The whole population studied comprised 539 patients (79% female, mean age at disease onset 55 years; RA and UA) whose allelic frequencies of the SNPs in TNFAIP3 are shown in Table 1. The minor allele (A) frequency (MAF) of rs17780429 was significantly lower in RA than in UA (13.4% vs 42.1%; p=0.018). In addition, the MAF (C) of rs600144 was lower in ACPA+ than in negatives patients (19.9% vs 38.6%; p=0.017). In the 235 patients who fulfilled the study criteria for response to MTX, it was observed that the presence of the T allele of rs11970361 showed a trend to lower response, assessed either with the EULAR or HUPI criteria, although statistical significance was not achieved, likely due to the low frequency of this allele. By contrast, carriers of the G allele of rs582757 were associated with lower HUPI response in an additive manner, reaching statistical significance for homozygous GG cases (OR 0.39 [0.16 to 0.91]; p=0.029).
Conclusion: Genetic variants of TNFAIP3 could explain phenotypic differences in early arthritis patients and might help to predict response to MTX. The low variability in the population of some of these SNPs would make difficult their implementation in clinical practice.
This study has been possible thanks to ISCIII grants PI18/00371 and PI21/0526, and unrestricted grant from Gebro Pharma.
Disclosures: A. Triguero, None; J. BALDIVIESO ACHA, None; E. Roy Vallejo, None; P. Quiroga-Colina, None; I. Llorente Cubas, None; R. Garcia-Vicuña, Novartis, Sanofi, Merck/MSD, Bristol-Myers Squibb(BMS), Eli Lilly, UCB, Janssen, Sandoz, Pfizer, Biogen; a. Puig kröger, None; I. Gonzalez, Gebro Pharma.
Background/Purpose: We recently described that methotrexate (MTX) could exert its immunomodulatory effect through the induction of A20 expression , a protein that negatively regulates NFkB signalling induced by TLR ligands or TNF stimulation. On the other hand, different single nucleotide polymorphisms (SNPs) in TNFAIP3, the gene that codes the A20 protein, have been associated with an increased risk to develop rheumatoid arthritis (RA). It is likely that these variants could be associated with lower expression and activity of A20.This study aims to determine whether those SNPs in TNFAIP3, associated with increased risk of RA, are useful for predicting MTX response.
Methods: We analysed data from PEARL study (Princesa Early Arthritis Register Longitudinal study; CEIC approval registry no 518; March 2011) in which sociodemographic, clinical, analytical, therapeutic variables and biological samples are collect by protocol in 5 structured visits.
We assessed clinical response (no, moderate and good) at six months in those patients who started MTX at the baseline and maintained this drug in monotherapy, by using the EULAR and HUPI response criteria. The following SNPs in TNFAIP3, rs600144, rs11970361, rs582757, rs11970411, and rs17780429, were genotyped using TaqMan probes (ThermoFisher Scientific; part number:C_7701163_10, C_238210_20, C_8300291_10, C_32241642_10, C_34723620_10, respectively).
To determine whether there was an association between the response to MTX and the SNPs, an order logistic regression model was performed using the ologit command of Stata 14.1. The model was adjusted for age, sex, MTX dose (median 15 mg; interquartile range 12.5 - 20) and disease activity at baseline.
Results: The whole population studied comprised 539 patients (79% female, mean age at disease onset 55 years; RA and UA) whose allelic frequencies of the SNPs in TNFAIP3 are shown in Table 1. The minor allele (A) frequency (MAF) of rs17780429 was significantly lower in RA than in UA (13.4% vs 42.1%; p=0.018). In addition, the MAF (C) of rs600144 was lower in ACPA+ than in negatives patients (19.9% vs 38.6%; p=0.017). In the 235 patients who fulfilled the study criteria for response to MTX, it was observed that the presence of the T allele of rs11970361 showed a trend to lower response, assessed either with the EULAR or HUPI criteria, although statistical significance was not achieved, likely due to the low frequency of this allele. By contrast, carriers of the G allele of rs582757 were associated with lower HUPI response in an additive manner, reaching statistical significance for homozygous GG cases (OR 0.39 [0.16 to 0.91]; p=0.029).
Conclusion: Genetic variants of TNFAIP3 could explain phenotypic differences in early arthritis patients and might help to predict response to MTX. The low variability in the population of some of these SNPs would make difficult their implementation in clinical practice.
This study has been possible thanks to ISCIII grants PI18/00371 and PI21/0526, and unrestricted grant from Gebro Pharma.

Disclosures: A. Triguero, None; J. BALDIVIESO ACHA, None; E. Roy Vallejo, None; P. Quiroga-Colina, None; I. Llorente Cubas, None; R. Garcia-Vicuña, Novartis, Sanofi, Merck/MSD, Bristol-Myers Squibb(BMS), Eli Lilly, UCB, Janssen, Sandoz, Pfizer, Biogen; a. Puig kröger, None; I. Gonzalez, Gebro Pharma.

