Back
Poster Session B
Systemic lupus erythematosus (SLE)
Session: (0629–0670) SLE – Etiology and Pathogenesis Poster
0645: Lupus Fibroblasts from Non-lesional Skin Exhibit Exaggerated Responses to Inflammatory Cytokines and Upregulate Pro-fibrotic Collagens in Patients with Scarring Lesions
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- SS
Suzanne Shoffner-Beck, BS
University of Michigan
Ann Arbor, MI, United States
Abstract Poster Presenter(s)
Suzanne Shoffner-Beck, Lisa Abernathy-Close, Stephanie Lazar, Amy Hurst, Craig Dobry, Deepika Pandian, Rachael Wasikowski, Kelly Arnold, Johann Gudjonsson, Lam Tsoi and J. Michelle Kahlenberg, University of Michigan, Ann Arbor, MI
Background/Purpose: Cutaneous lupus erythematosus (CLE) is a manifestation of systemic lupus erythematosus (SLE) that can cause significant patient distress and disfiguration secondary to scar. Scarring mechanisms remain poorly understood and interventions for preventing or treating scarring are lacking in SLE/CLE. Fibroblasts (FBs) are involved in the regulation of immune responses, inflammation, and scarring, yet the role of inflammatory mediators on FB function in SLE is unclear. Here, we examined the inflammatory phenotype in FBs isolated from non-lesional skin from healthy controls (HC), and SLE patients with and without scarring CLE.
Methods: Non-lesional punch biopsies were obtained from University of Michigan patients with SLE (n=21), divided into patients with scarring skin lesions (n=8) and non-scarring skin lesions (n=13), as well as HCs (n=34). Scarring disease status was determined using the damage score of the Cutaneous Lupus Erythematosus Disease Area and Severity Index (CLASI). FBs were isolated from punch biopsies, cultured, and stimulated with IFN-γ, IFN-ɑ, TNF-ɑ, TGF-𝛃, IL-1𝛃, or UV light. RNA sequencing was performed and differentially expressed genes (DEGs) were determined using DESeq2 with log2FC > 0.6 and adjusted p-value < 0.05. Pathway analysis was performed by filtering the genes with the largest effect size (log2FCCLE - log2FCHC) to identify pathways shared across all stimulations and unique to individual stimulations.
Results: HC and SLE FBs exhibited differential gene expression following cytokine treatment, with the largest effect size differences for TGF-𝛃, TNF-ɑ, IFN-γ, and IFN-ɑ stimulation. Cytokine-cytokine receptor signaling pathway genes were upregulated across all conditions, with relatively more upregulation in SLE FBs in comparison to healthy controls. DEGs included CXCL and CCL genes, interleukins, and TNF. (Fig 1)
Pathway analysis of the scarring and non-scarring states revealed DEGs in the IL-10 pathway, especially specific to stimulation with TGF-𝛃, a known fibrotic factor involved in myofibroblast activation. Interestingly, non-scarring patients had greater upregulation of proinflammatory signatures, including CXCL and CCL genes, and interleukins. Collagen trimer pathway genes were also significantly differentially expressed between scarring and non-scarring states. COL17A1 was up-regulated in scarring, especially after inflammatory signals like TGF-𝛃, TNF-ɑ, and IL-1𝛃, while COLQ, COL21A1, and COL4A3 were generally down-regulated in non-scarring. Histologic confirmation of collagen staining identified pro-fibrotic collagen expression in scarring skin lesions, near inflammatory infiltrates. (Fig 2)
Conclusion: Our results show that while exposure to inflammatory cytokines successfully up-regulates inflammatory pathways in both healthy and SLE fibroblasts, the effect is magnified in SLE. Furthermore, while all SLE fibroblasts are associated with elevated inflammation, FBs from patients who scar associated with upregulation of collagen pathways after TGF-β stimulation. Together, these findings provide important insights into the mechanisms of scarring in CLE and potential targets for intervention.
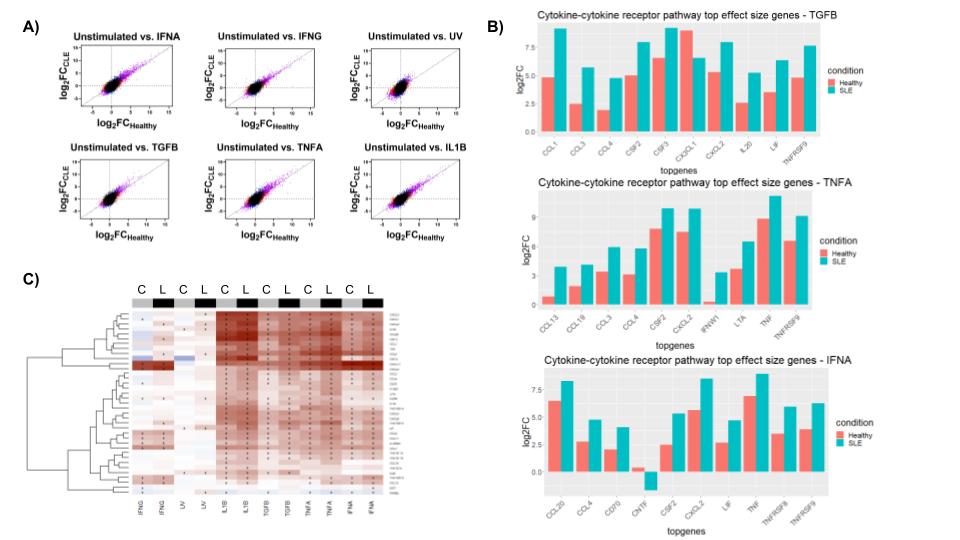 Figure 1: A) The effect sizes (log2FC) under indicated cytokine stimulations in healthy (x-axis) and lupus (y-axis) fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between SLE and healthy conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C) Heatmap illustrating the relatively higher upregulation of cytokine-cytokine receptor pathway genes in lupus compared to healthy controls.
Figure 1: A) The effect sizes (log2FC) under indicated cytokine stimulations in healthy (x-axis) and lupus (y-axis) fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between SLE and healthy conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C) Heatmap illustrating the relatively higher upregulation of cytokine-cytokine receptor pathway genes in lupus compared to healthy controls.
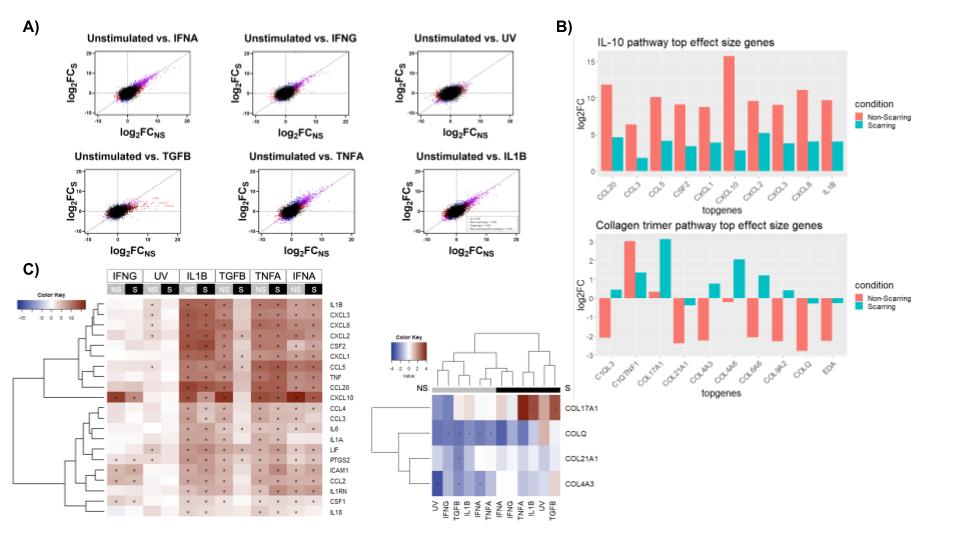 Figure 2: A) The effect sizes (log2FC) under indicated cytokine stimulations in non-scarring (x-axis) and scarring (y-axis) lupus fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between scarring and non-scarring conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C & D) Heatmaps illustrating the relatively higher upregulation of IL-10 pathway genes in non-scarring disease compared to scarring disease and the upregulation of collagen trimer pathway genes in scarring disease compared with non-scarring.
Figure 2: A) The effect sizes (log2FC) under indicated cytokine stimulations in non-scarring (x-axis) and scarring (y-axis) lupus fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between scarring and non-scarring conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C & D) Heatmaps illustrating the relatively higher upregulation of IL-10 pathway genes in non-scarring disease compared to scarring disease and the upregulation of collagen trimer pathway genes in scarring disease compared with non-scarring.
Disclosures: S. Shoffner-Beck, None; L. Abernathy-Close, None; S. Lazar, None; A. Hurst, None; C. Dobry, None; D. Pandian, None; R. Wasikowski, None; K. Arnold, None; J. Gudjonsson, None; L. Tsoi, None; J. Kahlenberg, AstraZeneca, Bristol-Myers Squibb(BMS), Eli Lilly, GlaxoSmithKlein(GSK), Janssen, Merck/MSD, Gilead.
Background/Purpose: Cutaneous lupus erythematosus (CLE) is a manifestation of systemic lupus erythematosus (SLE) that can cause significant patient distress and disfiguration secondary to scar. Scarring mechanisms remain poorly understood and interventions for preventing or treating scarring are lacking in SLE/CLE. Fibroblasts (FBs) are involved in the regulation of immune responses, inflammation, and scarring, yet the role of inflammatory mediators on FB function in SLE is unclear. Here, we examined the inflammatory phenotype in FBs isolated from non-lesional skin from healthy controls (HC), and SLE patients with and without scarring CLE.
Methods: Non-lesional punch biopsies were obtained from University of Michigan patients with SLE (n=21), divided into patients with scarring skin lesions (n=8) and non-scarring skin lesions (n=13), as well as HCs (n=34). Scarring disease status was determined using the damage score of the Cutaneous Lupus Erythematosus Disease Area and Severity Index (CLASI). FBs were isolated from punch biopsies, cultured, and stimulated with IFN-γ, IFN-ɑ, TNF-ɑ, TGF-𝛃, IL-1𝛃, or UV light. RNA sequencing was performed and differentially expressed genes (DEGs) were determined using DESeq2 with log2FC > 0.6 and adjusted p-value < 0.05. Pathway analysis was performed by filtering the genes with the largest effect size (log2FCCLE - log2FCHC) to identify pathways shared across all stimulations and unique to individual stimulations.
Results: HC and SLE FBs exhibited differential gene expression following cytokine treatment, with the largest effect size differences for TGF-𝛃, TNF-ɑ, IFN-γ, and IFN-ɑ stimulation. Cytokine-cytokine receptor signaling pathway genes were upregulated across all conditions, with relatively more upregulation in SLE FBs in comparison to healthy controls. DEGs included CXCL and CCL genes, interleukins, and TNF. (Fig 1)
Pathway analysis of the scarring and non-scarring states revealed DEGs in the IL-10 pathway, especially specific to stimulation with TGF-𝛃, a known fibrotic factor involved in myofibroblast activation. Interestingly, non-scarring patients had greater upregulation of proinflammatory signatures, including CXCL and CCL genes, and interleukins. Collagen trimer pathway genes were also significantly differentially expressed between scarring and non-scarring states. COL17A1 was up-regulated in scarring, especially after inflammatory signals like TGF-𝛃, TNF-ɑ, and IL-1𝛃, while COLQ, COL21A1, and COL4A3 were generally down-regulated in non-scarring. Histologic confirmation of collagen staining identified pro-fibrotic collagen expression in scarring skin lesions, near inflammatory infiltrates. (Fig 2)
Conclusion: Our results show that while exposure to inflammatory cytokines successfully up-regulates inflammatory pathways in both healthy and SLE fibroblasts, the effect is magnified in SLE. Furthermore, while all SLE fibroblasts are associated with elevated inflammation, FBs from patients who scar associated with upregulation of collagen pathways after TGF-β stimulation. Together, these findings provide important insights into the mechanisms of scarring in CLE and potential targets for intervention.
 Figure 1: A) The effect sizes (log2FC) under indicated cytokine stimulations in healthy (x-axis) and lupus (y-axis) fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between SLE and healthy conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C) Heatmap illustrating the relatively higher upregulation of cytokine-cytokine receptor pathway genes in lupus compared to healthy controls.
Figure 1: A) The effect sizes (log2FC) under indicated cytokine stimulations in healthy (x-axis) and lupus (y-axis) fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between SLE and healthy conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C) Heatmap illustrating the relatively higher upregulation of cytokine-cytokine receptor pathway genes in lupus compared to healthy controls. Figure 2: A) The effect sizes (log2FC) under indicated cytokine stimulations in non-scarring (x-axis) and scarring (y-axis) lupus fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between scarring and non-scarring conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C & D) Heatmaps illustrating the relatively higher upregulation of IL-10 pathway genes in non-scarring disease compared to scarring disease and the upregulation of collagen trimer pathway genes in scarring disease compared with non-scarring.
Figure 2: A) The effect sizes (log2FC) under indicated cytokine stimulations in non-scarring (x-axis) and scarring (y-axis) lupus fibroblasts. Diagonal lines represent what would be expected if there is no difference in gene expression between scarring and non-scarring conditions. B) Bar plots highlighting the differential gene expression for genes in pathways of interest. C & D) Heatmaps illustrating the relatively higher upregulation of IL-10 pathway genes in non-scarring disease compared to scarring disease and the upregulation of collagen trimer pathway genes in scarring disease compared with non-scarring.Disclosures: S. Shoffner-Beck, None; L. Abernathy-Close, None; S. Lazar, None; A. Hurst, None; C. Dobry, None; D. Pandian, None; R. Wasikowski, None; K. Arnold, None; J. Gudjonsson, None; L. Tsoi, None; J. Kahlenberg, AstraZeneca, Bristol-Myers Squibb(BMS), Eli Lilly, GlaxoSmithKlein(GSK), Janssen, Merck/MSD, Gilead.

