Back
Poster Session B
Rheumatoid arthritis (RA)
Session: (0589–0628) RA – Etiology and Pathogenesis Poster
0627: Spatial Transcriptomic Analysis Reveals Vascular Zonation of Myofibroblasts in Rheumatoid Arthritis Synovium
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- KB
Kartik Bhamidipati, PhD
Brigham and Women's Hospital
Boston, MA, United States
Abstract Poster Presenter(s)
Kartik Bhamidipati1, Roopa Madhu1, Sonia Presti1, Youngmi Kim2, Ye Cui2, Fan Zhang3, Anna Jonsson1, Aparna Nathan1, Nghia Millard1, Deepak Rao1, Laura Donlin4, Jennifer Anolik5, Soumya Raychaudhuri1, Michael Brenner6, Ilya Korsunsky1 and Kevin Wei7, 1Brigham and Women's Hospital, Boston, MA, 2Nanostring Technologies Inc., Seattle, WA, 3University of Colorado, Aurora, CO, 4Hospital for Special Surgery, New York, NY, 5University of Rochester Medical Center, Rochester, NY, 6Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 7Brigham and Women's Hospital and Harvard Medical School, Boston, MA
Background/Purpose: Many rheumatoid arthritis (RA) patients do not achieve sustained remission despite multiple lines of therapy. Recent studies have linked the expansion of synovial fibroblasts with treatment resistance in RA1. Using single-cell RNA-sequencing data generated by the Accelerating Medicines Partnership (AMP) Consortium, we identified the expansion of fibrotic myofibroblasts in RA distinguished by high expression of extracellular matrix proteins2. Here, we leveraged spatial transcriptomics and in vitro studies to reveal the vascular localization of myofibroblasts and identify the molecular drivers of this phenotype.
Methods: Spatial transcriptomic profiling: Synovial FFPE tissue sections (1 RA and 1 OA) were subject to cyclic in situ hybridization of 960 fluorescent mRNA probes and imaged using the CosMx™ Spatial Molecular Image (Nanostring). Cell segmentation was performed which was used to assign transcripts to cells. Subsequent clustering and visualization were performed.
Immunofluorescent staining: RA and OA synovial tissue sections were stained with antibodies against CD31, NOTCH3, POSTN, COL1A1, p-SMAD2/3 and TGF-β followed by secondary antibody staining and image acquisition.
Cell culture: Synovial fibroblasts were cultured and stimulated with Notch ligand or co-cultured with human umbilical vein endothelial cells (HUVECs). qPCR, ELISA, western blot, and flow cytometry of stimulated cells were performed.
Results: Synovial scRNAseq data generated from 79 donors revealed the significant expansion of POSTN+ fibrotic myofibroblasts, defined by high expression extracellular matrix proteins COL1A1 and COMP, in RA. To identify the localization of myofibroblasts within RA synovia we applied a myofibroblast gene signature to spatial transcriptomic data which showed localization of myofibroblasts to the perivascular niche. Immunofluorescent staining of myofibroblast markers POSTN and COL1A1 confirmed the perivascular deposition of these proteins in RA synovia. Further scRNAseq analysis revealed a myofibroblast differentiation trajectory encompassing Notch and TGF-β signaling. Stimulation of synovial fibroblasts with Notch ligand delta-like ligand 4 and co-culture with HUVECs strongly induced the expression of POSTN. Unexpectedly, we found that Notch signaling promoted strong expression of TGF-β in vitro. Immunofluorescent staining of synovial tissue confirmed perivascular localization of NOTCH3, TGF-β and pSMAD2/3, suggesting a molecular crosstalk between Notch and TGF-β during differentiation of myofibroblasts in RA synovia.
Conclusion: Our study leveraged cutting-edge spatial transcriptomics to identify endothelial cells as mediators of synovial tissue fibrosis via cellular crosstalk involving Notch and TGF-β signaling. Further investigation into the molecular basis of myofibroblast differentiation in RA synovia will enable the identification of novel therapies targeting myofibroblasts in treatment resistant RA.
1. Rivellese, F et al. Nat Med, 19 May. 2022, doi:10.1038/s41591-022-01789-0
2. Zhang, F. et al. bioRxiv 2022.02.25.481990 (2022) doi:10.1101/2022.02.25.481990.
.jpg) Figure 1: Expansion of the Myofibroblast Subset in RA
Figure 1: Expansion of the Myofibroblast Subset in RA
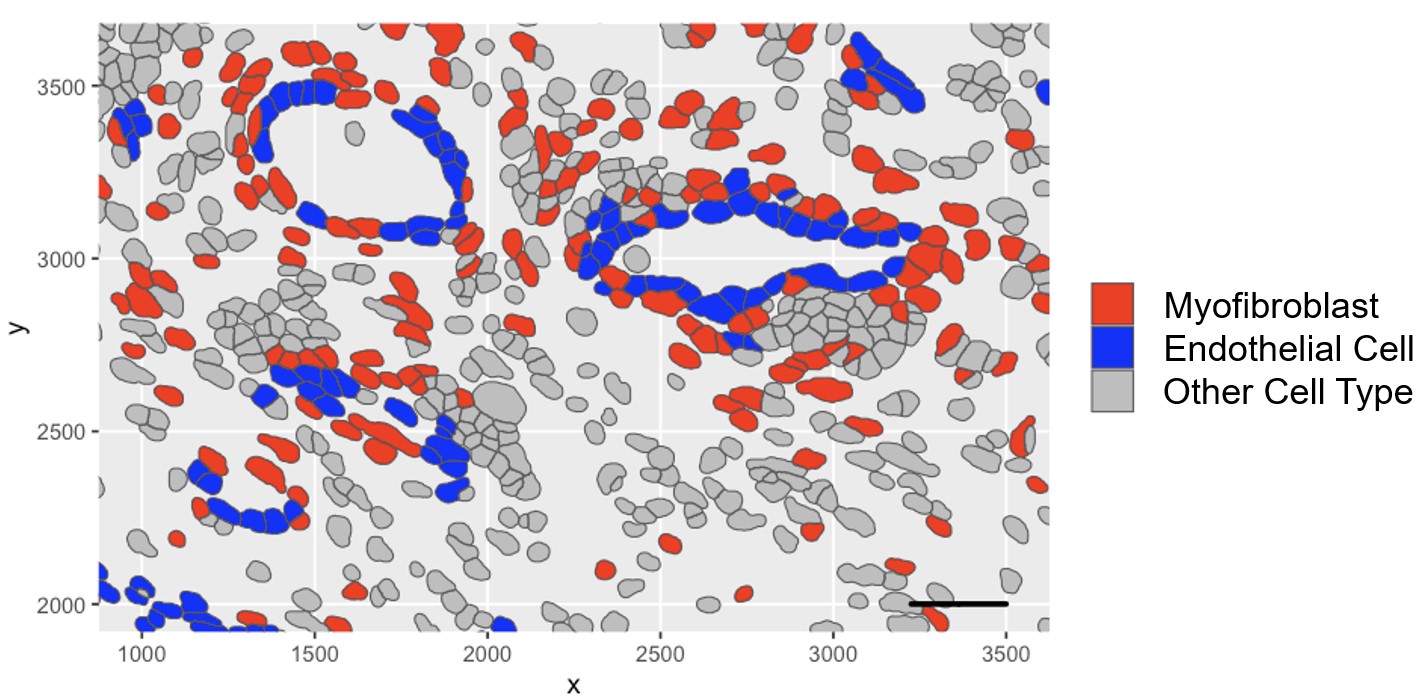 Figure 2: Spatial Transcriptomic Analysis Reveals Localization of Myofibroblasts to the Perivascular Niche
Figure 2: Spatial Transcriptomic Analysis Reveals Localization of Myofibroblasts to the Perivascular Niche
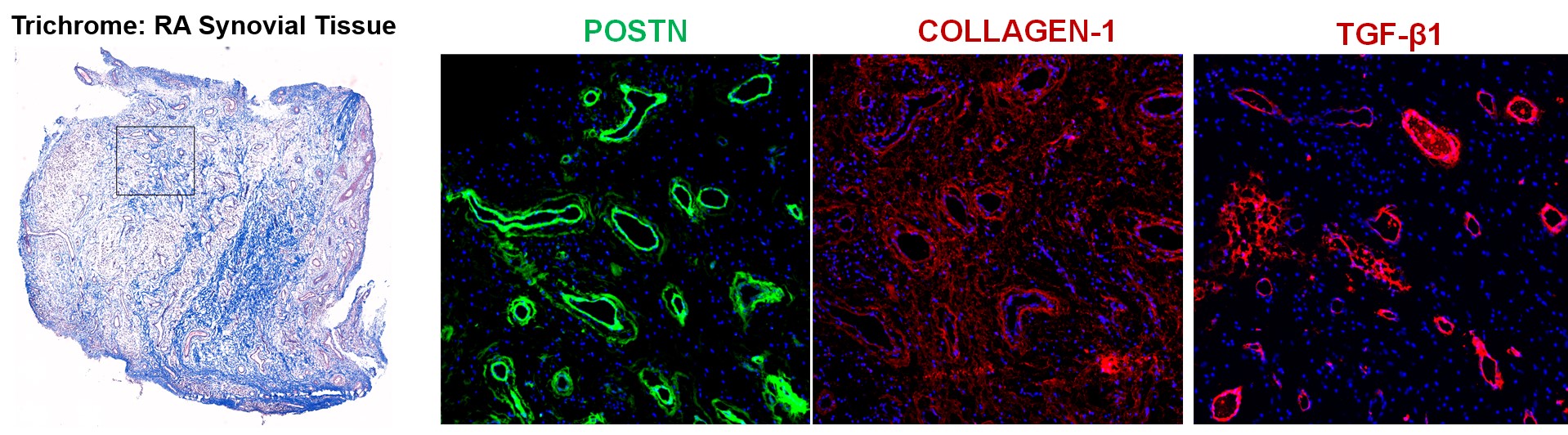 Figure 3: Extracellular Matrix Protein Deposition Occurs Perivascularly in RA Synovium
Figure 3: Extracellular Matrix Protein Deposition Occurs Perivascularly in RA Synovium
Disclosures: K. Bhamidipati, None; R. Madhu, None; S. Presti, None; Y. Kim, Nanostring Technologies; Y. Cui, Nanostring Technologies; F. Zhang, None; A. Jonsson, Amgen; A. Nathan, None; N. Millard, None; D. Rao, Janssen, Merck, Bristol-Myers Squibb, Scipher Medicine, HiFiBio, Inc., AstraZeneca, Pfizer; L. Donlin, None; J. Anolik, None; S. Raychaudhuri, Johnson & Johnson, Pfizer, Biogen, Gilead Sciences, Mestag Therapeutics; M. Brenner, GSK, 4FO Ventures, Mestag Therapeutics; I. Korsunsky, Mestag Therapeutics; K. Wei, Gilead sciences, Mestag, nanoString, 10X Genomics.
Background/Purpose: Many rheumatoid arthritis (RA) patients do not achieve sustained remission despite multiple lines of therapy. Recent studies have linked the expansion of synovial fibroblasts with treatment resistance in RA1. Using single-cell RNA-sequencing data generated by the Accelerating Medicines Partnership (AMP) Consortium, we identified the expansion of fibrotic myofibroblasts in RA distinguished by high expression of extracellular matrix proteins2. Here, we leveraged spatial transcriptomics and in vitro studies to reveal the vascular localization of myofibroblasts and identify the molecular drivers of this phenotype.
Methods: Spatial transcriptomic profiling: Synovial FFPE tissue sections (1 RA and 1 OA) were subject to cyclic in situ hybridization of 960 fluorescent mRNA probes and imaged using the CosMx™ Spatial Molecular Image (Nanostring). Cell segmentation was performed which was used to assign transcripts to cells. Subsequent clustering and visualization were performed.
Immunofluorescent staining: RA and OA synovial tissue sections were stained with antibodies against CD31, NOTCH3, POSTN, COL1A1, p-SMAD2/3 and TGF-β followed by secondary antibody staining and image acquisition.
Cell culture: Synovial fibroblasts were cultured and stimulated with Notch ligand or co-cultured with human umbilical vein endothelial cells (HUVECs). qPCR, ELISA, western blot, and flow cytometry of stimulated cells were performed.
Results: Synovial scRNAseq data generated from 79 donors revealed the significant expansion of POSTN+ fibrotic myofibroblasts, defined by high expression extracellular matrix proteins COL1A1 and COMP, in RA. To identify the localization of myofibroblasts within RA synovia we applied a myofibroblast gene signature to spatial transcriptomic data which showed localization of myofibroblasts to the perivascular niche. Immunofluorescent staining of myofibroblast markers POSTN and COL1A1 confirmed the perivascular deposition of these proteins in RA synovia. Further scRNAseq analysis revealed a myofibroblast differentiation trajectory encompassing Notch and TGF-β signaling. Stimulation of synovial fibroblasts with Notch ligand delta-like ligand 4 and co-culture with HUVECs strongly induced the expression of POSTN. Unexpectedly, we found that Notch signaling promoted strong expression of TGF-β in vitro. Immunofluorescent staining of synovial tissue confirmed perivascular localization of NOTCH3, TGF-β and pSMAD2/3, suggesting a molecular crosstalk between Notch and TGF-β during differentiation of myofibroblasts in RA synovia.
Conclusion: Our study leveraged cutting-edge spatial transcriptomics to identify endothelial cells as mediators of synovial tissue fibrosis via cellular crosstalk involving Notch and TGF-β signaling. Further investigation into the molecular basis of myofibroblast differentiation in RA synovia will enable the identification of novel therapies targeting myofibroblasts in treatment resistant RA.
1. Rivellese, F et al. Nat Med, 19 May. 2022, doi:10.1038/s41591-022-01789-0
2. Zhang, F. et al. bioRxiv 2022.02.25.481990 (2022) doi:10.1101/2022.02.25.481990.
.jpg) Figure 1: Expansion of the Myofibroblast Subset in RA
Figure 1: Expansion of the Myofibroblast Subset in RA Figure 2: Spatial Transcriptomic Analysis Reveals Localization of Myofibroblasts to the Perivascular Niche
Figure 2: Spatial Transcriptomic Analysis Reveals Localization of Myofibroblasts to the Perivascular Niche Figure 3: Extracellular Matrix Protein Deposition Occurs Perivascularly in RA Synovium
Figure 3: Extracellular Matrix Protein Deposition Occurs Perivascularly in RA SynoviumDisclosures: K. Bhamidipati, None; R. Madhu, None; S. Presti, None; Y. Kim, Nanostring Technologies; Y. Cui, Nanostring Technologies; F. Zhang, None; A. Jonsson, Amgen; A. Nathan, None; N. Millard, None; D. Rao, Janssen, Merck, Bristol-Myers Squibb, Scipher Medicine, HiFiBio, Inc., AstraZeneca, Pfizer; L. Donlin, None; J. Anolik, None; S. Raychaudhuri, Johnson & Johnson, Pfizer, Biogen, Gilead Sciences, Mestag Therapeutics; M. Brenner, GSK, 4FO Ventures, Mestag Therapeutics; I. Korsunsky, Mestag Therapeutics; K. Wei, Gilead sciences, Mestag, nanoString, 10X Genomics.

