Back
Plenary Session
Fibrosing rheumatic diseases (scleroderma, MCTD, IgG4-related disease, scleroderma mimics)
Session: Plenary II
1114: Computational Drug Repositioning and 3D Skin Models Identify EGFR and PI3K Inhibitors as Anti-fibrotic Targets for Systemic Sclerosis
Sunday, November 13, 2022
12:00 PM – 12:10 PM Eastern Time
Location: Exhibit Hall A
- DP
Dillon Popovich, BS
Dartmouth College
West Lebanon, NH, United States
Presenting Author(s)
Dillon Popovich1, Noelle Kosarek2, Rezvan Parvizi3, Tamar Abel4, Mengqi Huang5, Monica Espinoza6, Avi Smith7, Sasha Shenk7, Jonathan Garlick7 and Michael Whitfield8, 1Dartmouth College, West Lebanon, NH, 2Dartmouth Geisel School of Medicine, Lebanon, NH, 3Dartmouth, Lebanon, NH, 4Dartmouth College, Lebanon, NH, 5University of Pittsburgh, Pittsburgh, PN, 6Dartmouth College, Hanover, NH, 7Tufts University School of Dental Medicine, Boston, MA, 8Geisel School of Medicine, Lebanon, NH
Background/Purpose: Genome-wide analyses of gene expression in systemic sclerosis (SSc) skin biopsies have identified differential responses based on the mechanism of action of the therapy. The heterogeneity of SSc suggest that subset-specific gene expression could be used to reposition existing drugs for a personalized medicine approach to treatment. The goal of this study was to identify small molecules through the Connectivity Map 2.0 (CMAP) database which are predicted to reverse the gene expression changes observed in SSc skin and test the ability of these small molecules to attenuate fibrosis in a 3D skin-like model of SSc.
Methods: CMAP data was processed with RMA, quantile normalized, and fit to a multichip linear model. Probes were collapsed as average intensity and formatted for gene fold-change as the ratio of treatment to control intensities. DNA microarray data from Milano et al (GSE9285) was analyzed by Gene Set Variation Analysis (GSVA). Pathways statistically significant and specific to the inflammatory and fibroproliferative intrinsic subsets of SSc patients were determined. Small molecules from the CMAP data which regulated pathways specific for each SSc subset were identified and tested in our 3D tissue culture system, using fibroblasts derived from SSc patients or healthy individuals (Fig. 1A).
Results: Over 600 pathways were statistically significant in the fibroproliferative and inflammatory subsets of SSc patients using GSVA. Comparison to gene expression profiles in the CMAP database revealed EGFR inhibitors strongly downregulate pathways found in the inflammatory subset of SSc patients, while PI3K inhibitors were predicted to downregulate pathways in the fibroproliferative subset of SSc patients. Using self-assembled (SA) 3D skin-like tissues containing human dermal fibroblasts, we found no adverse effects on tissue growth with addition of inhibitor into tissue media. ECM deposition observed through H&E staining decreased while fibroblasts remained healthy. In SA tissues grown from SSc patient derived fibroblasts, thickness decreased significantly after treatment with either a PI3K or EGFR inhibitor. SA tissues using fibroblasts derived from healthy individuals showed no significant changes in tissue thickness and resembled tissues previously cultured from commercially available human dermal fibroblasts (Fig. 1 B, C). Immunohistochemical staining shows EGFR overexpression in SSc compared to healthy tissues. PI3K pathway upregulation has been shown in prior analyses to be upregulated in patients with SSc. Western blotting and qPCR were performed to determine SSc-specific overactivation of the pathway in cell culture as well as SA tissues.
Conclusion: We identified multiple small molecule inhibitors that downregulate the gene expression pathways shown to be highly expressed in SSc skin biopsies. Experimental validation in 3D skin-like tissues constructed from SSc or healthy dermal fibroblasts showed a marked specific decrease in SSc tissue thickness, suggesting reduced ECM deposition upon exposure.
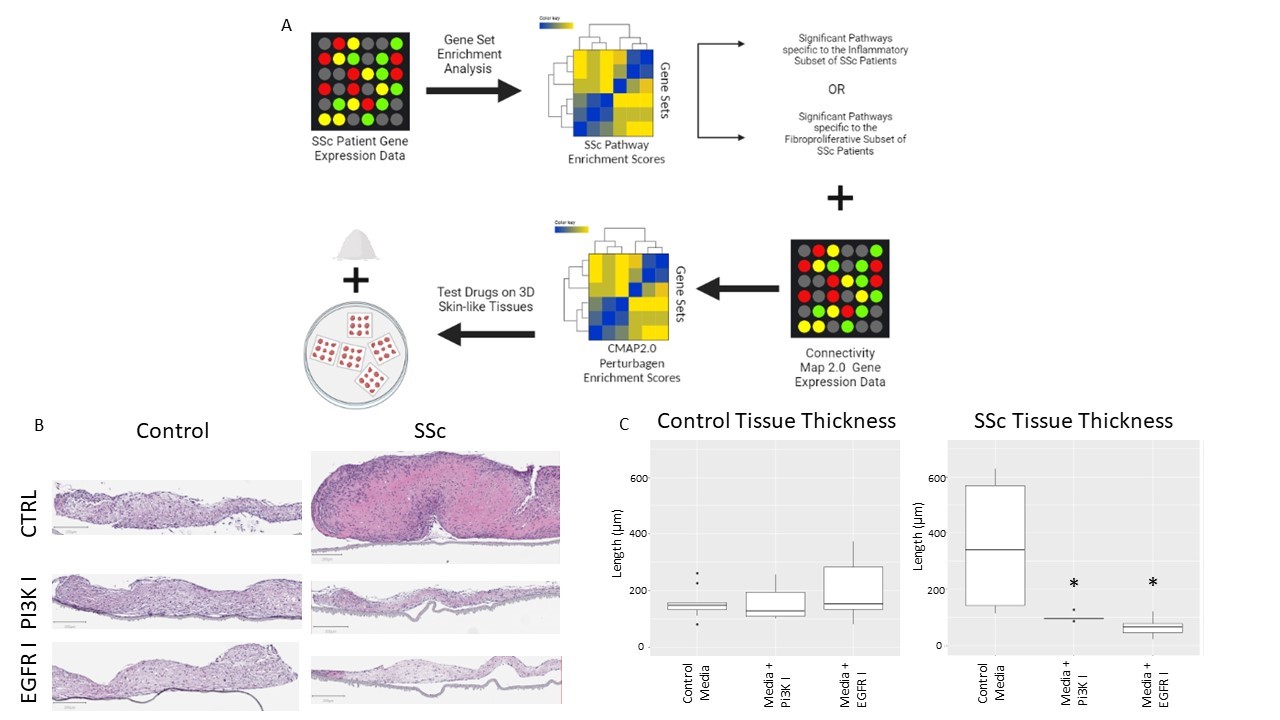 Figure 1. A) General outline of drug repositioning pipeline. GSVA is run on microarray datasets for SSc patient cohorts. Significant gene sets for each gene expression subset are then analyzed in CMAP data to find drugs which regulate these pathways. Drugs found to heavily offset this gene set dysregulation were then used in our 3D skin-like tissue culture to determine drug specific effects. B) H&E staining of healthy and SSc derived self-assembled tissues treated with growth media as control (CTRL), growth media added with PI3K inhibitor (PI3K I), or EGFR inhibitor (EGFR I). C) Tissue thickness measurements of tissues harvested in B (including replicates). * = p < 0.05
Figure 1. A) General outline of drug repositioning pipeline. GSVA is run on microarray datasets for SSc patient cohorts. Significant gene sets for each gene expression subset are then analyzed in CMAP data to find drugs which regulate these pathways. Drugs found to heavily offset this gene set dysregulation were then used in our 3D skin-like tissue culture to determine drug specific effects. B) H&E staining of healthy and SSc derived self-assembled tissues treated with growth media as control (CTRL), growth media added with PI3K inhibitor (PI3K I), or EGFR inhibitor (EGFR I). C) Tissue thickness measurements of tissues harvested in B (including replicates). * = p < 0.05
Disclosures: D. Popovich, None; N. Kosarek, None; R. Parvizi, None; T. Abel, None; M. Huang, None; M. Espinoza, None; A. Smith, None; S. Shenk, None; J. Garlick, None; M. Whitfield, Bristol-Myers Squibb(BMS), Celdara Medical LLC.
Background/Purpose: Genome-wide analyses of gene expression in systemic sclerosis (SSc) skin biopsies have identified differential responses based on the mechanism of action of the therapy. The heterogeneity of SSc suggest that subset-specific gene expression could be used to reposition existing drugs for a personalized medicine approach to treatment. The goal of this study was to identify small molecules through the Connectivity Map 2.0 (CMAP) database which are predicted to reverse the gene expression changes observed in SSc skin and test the ability of these small molecules to attenuate fibrosis in a 3D skin-like model of SSc.
Methods: CMAP data was processed with RMA, quantile normalized, and fit to a multichip linear model. Probes were collapsed as average intensity and formatted for gene fold-change as the ratio of treatment to control intensities. DNA microarray data from Milano et al (GSE9285) was analyzed by Gene Set Variation Analysis (GSVA). Pathways statistically significant and specific to the inflammatory and fibroproliferative intrinsic subsets of SSc patients were determined. Small molecules from the CMAP data which regulated pathways specific for each SSc subset were identified and tested in our 3D tissue culture system, using fibroblasts derived from SSc patients or healthy individuals (Fig. 1A).
Results: Over 600 pathways were statistically significant in the fibroproliferative and inflammatory subsets of SSc patients using GSVA. Comparison to gene expression profiles in the CMAP database revealed EGFR inhibitors strongly downregulate pathways found in the inflammatory subset of SSc patients, while PI3K inhibitors were predicted to downregulate pathways in the fibroproliferative subset of SSc patients. Using self-assembled (SA) 3D skin-like tissues containing human dermal fibroblasts, we found no adverse effects on tissue growth with addition of inhibitor into tissue media. ECM deposition observed through H&E staining decreased while fibroblasts remained healthy. In SA tissues grown from SSc patient derived fibroblasts, thickness decreased significantly after treatment with either a PI3K or EGFR inhibitor. SA tissues using fibroblasts derived from healthy individuals showed no significant changes in tissue thickness and resembled tissues previously cultured from commercially available human dermal fibroblasts (Fig. 1 B, C). Immunohistochemical staining shows EGFR overexpression in SSc compared to healthy tissues. PI3K pathway upregulation has been shown in prior analyses to be upregulated in patients with SSc. Western blotting and qPCR were performed to determine SSc-specific overactivation of the pathway in cell culture as well as SA tissues.
Conclusion: We identified multiple small molecule inhibitors that downregulate the gene expression pathways shown to be highly expressed in SSc skin biopsies. Experimental validation in 3D skin-like tissues constructed from SSc or healthy dermal fibroblasts showed a marked specific decrease in SSc tissue thickness, suggesting reduced ECM deposition upon exposure.
 Figure 1. A) General outline of drug repositioning pipeline. GSVA is run on microarray datasets for SSc patient cohorts. Significant gene sets for each gene expression subset are then analyzed in CMAP data to find drugs which regulate these pathways. Drugs found to heavily offset this gene set dysregulation were then used in our 3D skin-like tissue culture to determine drug specific effects. B) H&E staining of healthy and SSc derived self-assembled tissues treated with growth media as control (CTRL), growth media added with PI3K inhibitor (PI3K I), or EGFR inhibitor (EGFR I). C) Tissue thickness measurements of tissues harvested in B (including replicates). * = p < 0.05
Figure 1. A) General outline of drug repositioning pipeline. GSVA is run on microarray datasets for SSc patient cohorts. Significant gene sets for each gene expression subset are then analyzed in CMAP data to find drugs which regulate these pathways. Drugs found to heavily offset this gene set dysregulation were then used in our 3D skin-like tissue culture to determine drug specific effects. B) H&E staining of healthy and SSc derived self-assembled tissues treated with growth media as control (CTRL), growth media added with PI3K inhibitor (PI3K I), or EGFR inhibitor (EGFR I). C) Tissue thickness measurements of tissues harvested in B (including replicates). * = p < 0.05Disclosures: D. Popovich, None; N. Kosarek, None; R. Parvizi, None; T. Abel, None; M. Huang, None; M. Espinoza, None; A. Smith, None; S. Shenk, None; J. Garlick, None; M. Whitfield, Bristol-Myers Squibb(BMS), Celdara Medical LLC.

