Back
Poster Session D
Osteoarthritis (OA) and related disorders
Session: (1888–1923) Osteoarthritis – Clinical Poster
1895: Association Between Synovial Tissue Metabolomic Profile and Pain in Patients with Knee Osteoarthritis
Monday, November 14, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall

Jessica Murillo Saich, PhD, BSc
University of California San Diego
La Jolla, CA, United States
Abstract Poster Presenter(s)
Jessica Murillo Saich1, Roxana Coras2, Robert Meyer3, Nancy Lane4 and Monica Guma5, 1University of California San Diego, La Jolla, CA, 2University of California San Diego, San Diego, CA, 3San Diego VA Healthcare Service, San Diego, CA, 4University of California Davis, Hillsborough, CA, 5UCSD, La Jolla, CA
Background/Purpose: Increasing evidence indicates that osteoarthritis (OA) progression is mediated by low-grade synovial inflammation (synovitis) that contributes to joint pain and stiffness, especially in knee OA (KOA). In addition, chronic inflammatory responses are associated with dramatic shifts in tissue metabolism. The objective of this work is to explore the metabolomic profiling in synovial tissue (ST) from OA patients and its association with pain.
Methods: Synovial tissue (ST) and synovial fluid (SF) samples were collected from patients with OA during knee replacement. ST was snap-frozen for metabolomic analysis. The patients were interrogated about nociceptive and neuropathic pain using WOMAC and the painDETECT questionnaires respectively. Nerve Growth Factor (NGF) was quantified in SF by enzyme-linked immunosorbent assay (ELISA). A 600 MHz Bruker Avance III spectrometer 1H-NMR was used to acquire NMR spectra of ST samples. Software Chenomx NMR suite 8.5 professional was used for metabolite identification and quantification. The samples were normalized by volume/weight and the concentrations were reported in μM. Metaboanalyst 5.0, SPSS v26, and R studio were used for statistical analysis.
Results: 23 male patients (average age: 68 years, standard deviation (SD) 6.59, average BMI 28.2, SD 3.9) were recruited. At the time of the surgery, patients had a mean total WOMAC of 54.5 (SD, 18.2), WOMAC pain of 11.63 (SD 13.55), WOMAC stiffness of 4.7 (SD 1.6), WOMAC difficulty of 37.9 (SD 13.5), and painDETECT of 14.9 (SD 7.5). NGF levels in SF had a mean of 34 (SD 14.5) ng/mL. 47.8% of patients were classified as low pain (WOMAC pain ≤10) and 52.2% as high pain (WOMAC pain >10). For neuropathic pain, 37.5% were classified as unlikely (PainDETECT ≤12), and 62.5% likely (PainDETECT >12). NGF from SF was positive correlated with painDETECT (r=0.60, p=0.030) but not with WOMAC pain (r=0.21, p=0.35). Several metabolites, including creatine phosphate, glycine and tyrosine had a positive correlation with painDETECT, and acetoacetate had a negative correlation. Dimethylamine, proline, tryptophan and valine correlated positively and threonine negatively with WOMAC pain. The orthogonal partial least square discriminant analysis (OPLS/DA) showed a good separation of patients for painDETECT and WOMAC pain Phenylalanine and tyrosine metabolism, and phenylalanine, tyrosine and tryptophan biosynthesis were enriched metabolic pathways in patients with likely neuropathic pain (Figure 1), while pirimidine and tryptophan metabolism contributed to the discrimination between low and high WOMAC pain (Figure 2).
Conclusion: Neuropathic and nociceptive pain showed a different association with metabolomic profiling in tissue with OA. NGF in SF was associated only to neuropathic pain. Several metabolites differentially correlated with WOMAC and painDETECT suggesting different metabolic pathways involved in knee OA pain. Further studies are needed to determine whether metabolomic profiling of synovial tissue can identify new therapeutic targets for pain of OA.
.jpg) Figure 1. Metabolomic profiling discriminates patients with likely and unlikely neuropathic pain. A) OPLS-DA according to painDETECT classification showed a clear separation between groups with good predictive performance (Q2=0.102); B) Phenylalanine, tyrosine and serine were the 3 metabolites with higher VIP score; C) Enrichment pathways showed that phenylalanine and tyrosine metabolism are higher in patients with likely neurpatic pain according to painDETECT.
Figure 1. Metabolomic profiling discriminates patients with likely and unlikely neuropathic pain. A) OPLS-DA according to painDETECT classification showed a clear separation between groups with good predictive performance (Q2=0.102); B) Phenylalanine, tyrosine and serine were the 3 metabolites with higher VIP score; C) Enrichment pathways showed that phenylalanine and tyrosine metabolism are higher in patients with likely neurpatic pain according to painDETECT.
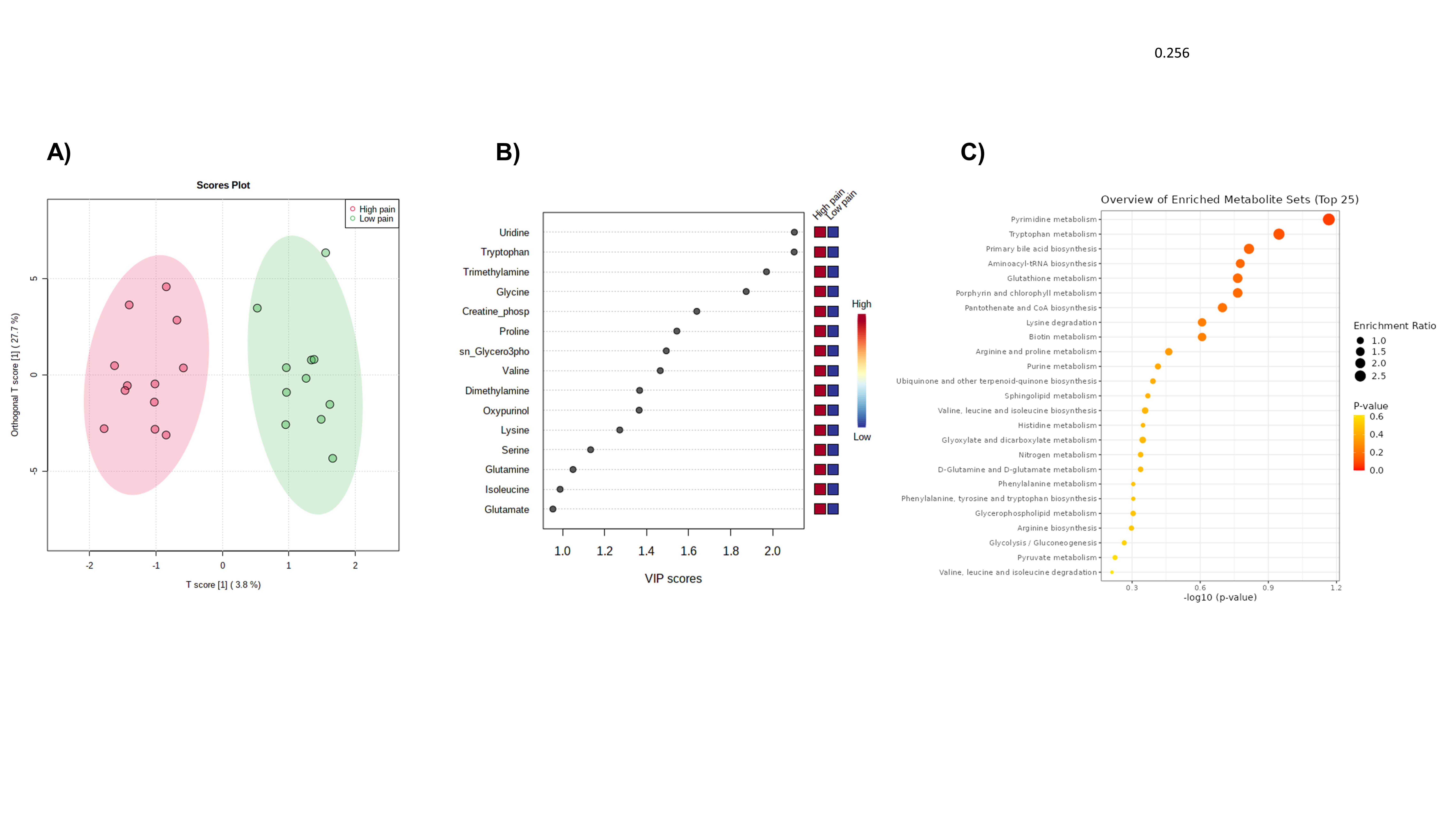 Figure 2. Metabolomic profiling discriminates patients with high and low nocioceptive pain. A) OPLS-DA between low pain versus high WOMAC pain shows a clear separation between groups with good predictive performance (Q2=0.256); B) Uridine, tryptophan and trimethylamine were the metabolites with higher VIP score; C) Enrichment pathways showed that pirimidine and tryptophan metabolism contribute to the discrimination between low and high WOMAC pain
Figure 2. Metabolomic profiling discriminates patients with high and low nocioceptive pain. A) OPLS-DA between low pain versus high WOMAC pain shows a clear separation between groups with good predictive performance (Q2=0.256); B) Uridine, tryptophan and trimethylamine were the metabolites with higher VIP score; C) Enrichment pathways showed that pirimidine and tryptophan metabolism contribute to the discrimination between low and high WOMAC pain
Disclosures: J. Murillo Saich, None; R. Coras, None; R. Meyer, None; N. Lane, Amgen, GlaxoSmithKlein(GSK); M. Guma, Pfizer, Novartis, Gilead, Sonoma Bio, Genentech.
Background/Purpose: Increasing evidence indicates that osteoarthritis (OA) progression is mediated by low-grade synovial inflammation (synovitis) that contributes to joint pain and stiffness, especially in knee OA (KOA). In addition, chronic inflammatory responses are associated with dramatic shifts in tissue metabolism. The objective of this work is to explore the metabolomic profiling in synovial tissue (ST) from OA patients and its association with pain.
Methods: Synovial tissue (ST) and synovial fluid (SF) samples were collected from patients with OA during knee replacement. ST was snap-frozen for metabolomic analysis. The patients were interrogated about nociceptive and neuropathic pain using WOMAC and the painDETECT questionnaires respectively. Nerve Growth Factor (NGF) was quantified in SF by enzyme-linked immunosorbent assay (ELISA). A 600 MHz Bruker Avance III spectrometer 1H-NMR was used to acquire NMR spectra of ST samples. Software Chenomx NMR suite 8.5 professional was used for metabolite identification and quantification. The samples were normalized by volume/weight and the concentrations were reported in μM. Metaboanalyst 5.0, SPSS v26, and R studio were used for statistical analysis.
Results: 23 male patients (average age: 68 years, standard deviation (SD) 6.59, average BMI 28.2, SD 3.9) were recruited. At the time of the surgery, patients had a mean total WOMAC of 54.5 (SD, 18.2), WOMAC pain of 11.63 (SD 13.55), WOMAC stiffness of 4.7 (SD 1.6), WOMAC difficulty of 37.9 (SD 13.5), and painDETECT of 14.9 (SD 7.5). NGF levels in SF had a mean of 34 (SD 14.5) ng/mL. 47.8% of patients were classified as low pain (WOMAC pain ≤10) and 52.2% as high pain (WOMAC pain >10). For neuropathic pain, 37.5% were classified as unlikely (PainDETECT ≤12), and 62.5% likely (PainDETECT >12). NGF from SF was positive correlated with painDETECT (r=0.60, p=0.030) but not with WOMAC pain (r=0.21, p=0.35). Several metabolites, including creatine phosphate, glycine and tyrosine had a positive correlation with painDETECT, and acetoacetate had a negative correlation. Dimethylamine, proline, tryptophan and valine correlated positively and threonine negatively with WOMAC pain. The orthogonal partial least square discriminant analysis (OPLS/DA) showed a good separation of patients for painDETECT and WOMAC pain Phenylalanine and tyrosine metabolism, and phenylalanine, tyrosine and tryptophan biosynthesis were enriched metabolic pathways in patients with likely neuropathic pain (Figure 1), while pirimidine and tryptophan metabolism contributed to the discrimination between low and high WOMAC pain (Figure 2).
Conclusion: Neuropathic and nociceptive pain showed a different association with metabolomic profiling in tissue with OA. NGF in SF was associated only to neuropathic pain. Several metabolites differentially correlated with WOMAC and painDETECT suggesting different metabolic pathways involved in knee OA pain. Further studies are needed to determine whether metabolomic profiling of synovial tissue can identify new therapeutic targets for pain of OA.
.jpg) Figure 1. Metabolomic profiling discriminates patients with likely and unlikely neuropathic pain. A) OPLS-DA according to painDETECT classification showed a clear separation between groups with good predictive performance (Q2=0.102); B) Phenylalanine, tyrosine and serine were the 3 metabolites with higher VIP score; C) Enrichment pathways showed that phenylalanine and tyrosine metabolism are higher in patients with likely neurpatic pain according to painDETECT.
Figure 1. Metabolomic profiling discriminates patients with likely and unlikely neuropathic pain. A) OPLS-DA according to painDETECT classification showed a clear separation between groups with good predictive performance (Q2=0.102); B) Phenylalanine, tyrosine and serine were the 3 metabolites with higher VIP score; C) Enrichment pathways showed that phenylalanine and tyrosine metabolism are higher in patients with likely neurpatic pain according to painDETECT. Figure 2. Metabolomic profiling discriminates patients with high and low nocioceptive pain. A) OPLS-DA between low pain versus high WOMAC pain shows a clear separation between groups with good predictive performance (Q2=0.256); B) Uridine, tryptophan and trimethylamine were the metabolites with higher VIP score; C) Enrichment pathways showed that pirimidine and tryptophan metabolism contribute to the discrimination between low and high WOMAC pain
Figure 2. Metabolomic profiling discriminates patients with high and low nocioceptive pain. A) OPLS-DA between low pain versus high WOMAC pain shows a clear separation between groups with good predictive performance (Q2=0.256); B) Uridine, tryptophan and trimethylamine were the metabolites with higher VIP score; C) Enrichment pathways showed that pirimidine and tryptophan metabolism contribute to the discrimination between low and high WOMAC painDisclosures: J. Murillo Saich, None; R. Coras, None; R. Meyer, None; N. Lane, Amgen, GlaxoSmithKlein(GSK); M. Guma, Pfizer, Novartis, Gilead, Sonoma Bio, Genentech.

