Back
Poster Session A
Pediatric autoimmune diseases: Kawasaki disease, juvenile dermatomyositis and juvenile localized scleroderma
Session: (0034–0044) Pediatric Rheumatology – Basic Science Poster
0040: Plasma Proteomic Signatures in Juvenile Dermatomyositis Highlight Novel Proteins and a Protein Module That Associate with Skin and Global Disease Activity
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- MK
Marianne Kerski, MD
University of Michigan
Ann Arbor, MI, United States
Abstract Poster Presenter(s)
Marianne Kerski1, Celine Berthier1, Alex Tsoi1, Sarah Vandenbergen1, Madison McClune1, Corey Powell1, J. Michelle Kahlenberg1 and Jessica Turnier2, 1University of Michigan, Ann Arbor, MI, 2University of Michigan, Saline, MI
Background/Purpose: Juvenile dermatomyositis (JDM) patients display heterogenous disease phenotypes and treatment response patterns. Our study used a multiplexed proteomics assay for measurement of 3072 proteins in plasma of a JDM cohort. The objectives were to (1) identify differentially regulated proteins (DRP) in JDM compared to healthy controls (CTL), (2) utilize weighted gene co-expression network analysis (WCGNA) to identify protein modules and (3) determine the association of module-associated proteins with disease activity measures.
Methods: We measured plasma protein levels using the Olink® Explore 3072 panel in a cross-sectional cohort of 32 JDM patients meeting 2017 EULAR/ACR classification criteria and 13 CTL. We phenotyped patients using the Cutaneous Dermatomyositis Disease Area and Severity Index (CDASI), Manual Muscle Testing (MMT-8), Childhood Myositis Assessment Scale (CMAS), and Physician's Global Assessment of Disease Activity (PGA). We performed differential protein expression analysis (false discovery rate, FDR< 0.10) and used linear regression models, adjusting for age, gender, disease activity, and treatment status (p < 0.05). WGCNA was performed to identify protein modules; module protein expression z-scores were correlated with clinical disease activity measures (Pearson correlation, p < 0.05). Ingenuity Pathway Analysis (IPA) was performed to identify biological pathways regulated (p< 0.05) for proteins and modules.
Results: We identified 229 and 20 DRP in JDM compared to CTL using linear regression and differential protein expression analysis respectively. Top regulated pathways represented by these DRP included STAT3, death receptor signaling, and granulocyte adhesion and diapedesis (p< 0.001). Treatment naïve JDM as compared to CTL and active JDM on treatment identified 63 and 40 DRP respectively, including CXCL10, MYOM3, IL10, IDO1, MYBPC1, NOS1 and KLHL41 (Figure 1). PGA, CDASI, MMT-8, and CMAS associated with 70, 106, 8, and 2 proteins, respectively (Figure 2). Top proteins associating with PGA were CXCL10 (r=0.84), CCL7 (r=0.78), MYL3 (r=0.76), RBFOX3 (r=0.75), and CAPN3 (r=0.71) (Figure 2). CDASI most strongly associated with CCL7 (r=0.77), KLHL41 (r=0.77), TNFRSF6B (r=0.73), WARS (r=0.73), and CXCL10 (r=0.72) (Figure 2). WCGNA identified five modules, and module 4 (n=417 proteins) expression scores associated with CDASI (r=0.44, p=0.01), PGA (r=0.38, p=0.03) and serum muscle enzyme levels. 63 proteins included in module 4 associated with at least one disease activity measure. Top upstream regulators in module 4 (p< 0.001) were IL-1B, IL-6, and TNF. Biological pathways regulated in module 4 included acute phase response signaling, coagulation system, and complement system (p < 0.001) (Figure 3).
Conclusion: We identified novel DRP in JDM involved in metabolic dysregulation (CAPN3 and IDO1), IFN signaling (CXCL10) and muscle structure and myogenesis. WCGNA identified a protein module associated with JDM skin and global disease activity enriched for proteins involved in acute phase response signaling and complement system. Further study of candidate proteins may assist in development of personalized care for JDM patients.
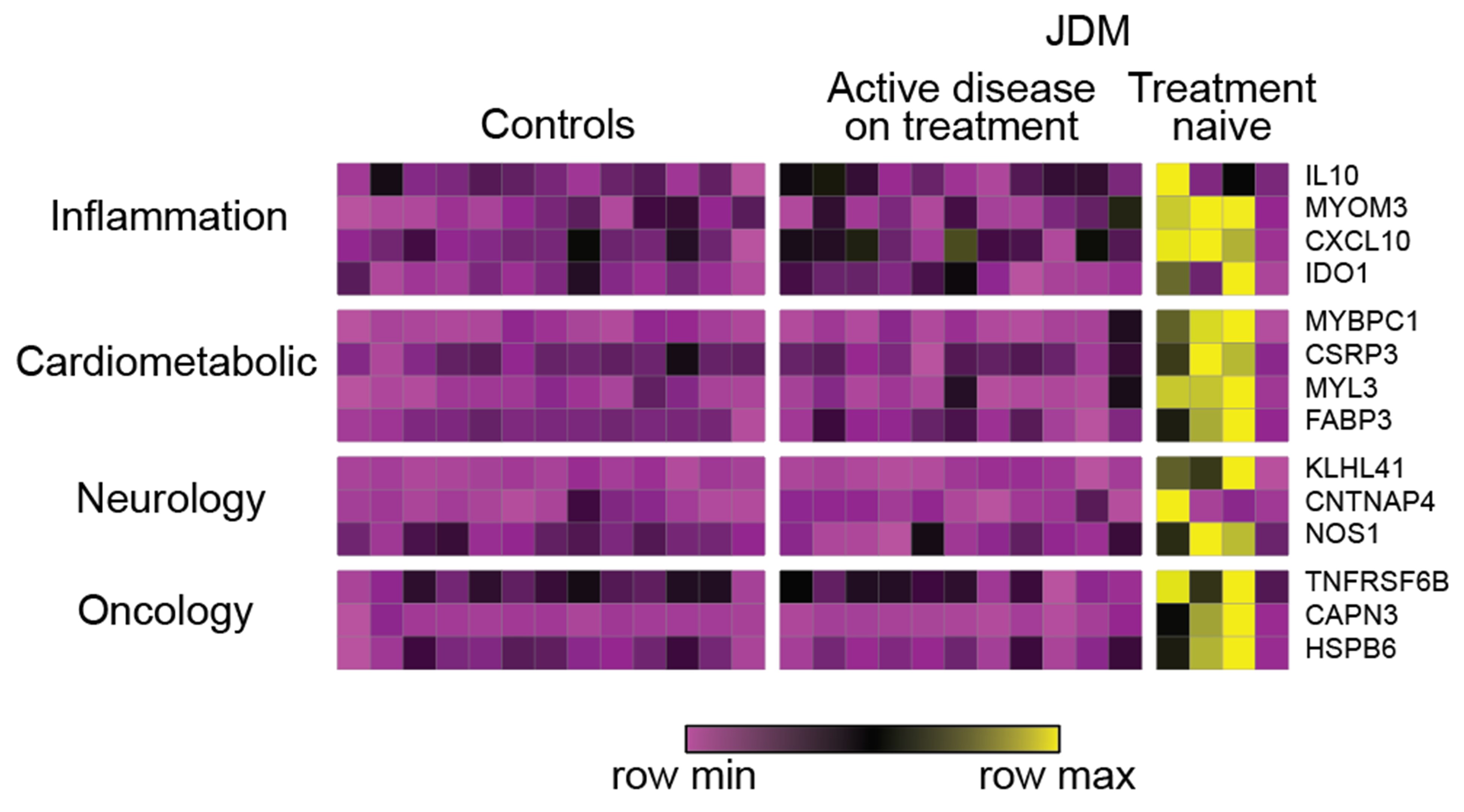 Heat map of differentially regulated proteins in treatment naïve JDM patients relative to active JDM on treatment and CTL (FDR < 0.10). Each column represents an individual patient sample while each row represents a differentially expressed protein in treatment naïve JDM relative to CTL and active disease on treatment. Included DRP were those with greatest fold change from each sub-panel from the Olink® Explore 3072 panel. Protein expression values are depicted using the color scale shown with purple to yellow indicating increasing expression.
Heat map of differentially regulated proteins in treatment naïve JDM patients relative to active JDM on treatment and CTL (FDR < 0.10). Each column represents an individual patient sample while each row represents a differentially expressed protein in treatment naïve JDM relative to CTL and active disease on treatment. Included DRP were those with greatest fold change from each sub-panel from the Olink® Explore 3072 panel. Protein expression values are depicted using the color scale shown with purple to yellow indicating increasing expression.
.jpg) Correlation of candidate protein biomarkers with PGA and CDASI (p-value < 0.001)
Correlation of candidate protein biomarkers with PGA and CDASI (p-value < 0.001)
.jpg) Predominant regulated pathways from proteins included in module 4 identified using WGCNA. The dot size represents the number of proteins regulated in each pathway compared to the total number of proteins in the pathway (protein ratio). The dot color represents the -log(p-value).
Predominant regulated pathways from proteins included in module 4 identified using WGCNA. The dot size represents the number of proteins regulated in each pathway compared to the total number of proteins in the pathway (protein ratio). The dot color represents the -log(p-value).
Disclosures: M. Kerski, None; C. Berthier, None; A. Tsoi, None; S. Vandenbergen, None; M. McClune, None; C. Powell, None; J. Kahlenberg, AstraZeneca, Bristol-Myers Squibb(BMS), Eli Lilly, GlaxoSmithKlein(GSK), Janssen, Merck/MSD, Gilead; J. Turnier, None.
Background/Purpose: Juvenile dermatomyositis (JDM) patients display heterogenous disease phenotypes and treatment response patterns. Our study used a multiplexed proteomics assay for measurement of 3072 proteins in plasma of a JDM cohort. The objectives were to (1) identify differentially regulated proteins (DRP) in JDM compared to healthy controls (CTL), (2) utilize weighted gene co-expression network analysis (WCGNA) to identify protein modules and (3) determine the association of module-associated proteins with disease activity measures.
Methods: We measured plasma protein levels using the Olink® Explore 3072 panel in a cross-sectional cohort of 32 JDM patients meeting 2017 EULAR/ACR classification criteria and 13 CTL. We phenotyped patients using the Cutaneous Dermatomyositis Disease Area and Severity Index (CDASI), Manual Muscle Testing (MMT-8), Childhood Myositis Assessment Scale (CMAS), and Physician's Global Assessment of Disease Activity (PGA). We performed differential protein expression analysis (false discovery rate, FDR< 0.10) and used linear regression models, adjusting for age, gender, disease activity, and treatment status (p < 0.05). WGCNA was performed to identify protein modules; module protein expression z-scores were correlated with clinical disease activity measures (Pearson correlation, p < 0.05). Ingenuity Pathway Analysis (IPA) was performed to identify biological pathways regulated (p< 0.05) for proteins and modules.
Results: We identified 229 and 20 DRP in JDM compared to CTL using linear regression and differential protein expression analysis respectively. Top regulated pathways represented by these DRP included STAT3, death receptor signaling, and granulocyte adhesion and diapedesis (p< 0.001). Treatment naïve JDM as compared to CTL and active JDM on treatment identified 63 and 40 DRP respectively, including CXCL10, MYOM3, IL10, IDO1, MYBPC1, NOS1 and KLHL41 (Figure 1). PGA, CDASI, MMT-8, and CMAS associated with 70, 106, 8, and 2 proteins, respectively (Figure 2). Top proteins associating with PGA were CXCL10 (r=0.84), CCL7 (r=0.78), MYL3 (r=0.76), RBFOX3 (r=0.75), and CAPN3 (r=0.71) (Figure 2). CDASI most strongly associated with CCL7 (r=0.77), KLHL41 (r=0.77), TNFRSF6B (r=0.73), WARS (r=0.73), and CXCL10 (r=0.72) (Figure 2). WCGNA identified five modules, and module 4 (n=417 proteins) expression scores associated with CDASI (r=0.44, p=0.01), PGA (r=0.38, p=0.03) and serum muscle enzyme levels. 63 proteins included in module 4 associated with at least one disease activity measure. Top upstream regulators in module 4 (p< 0.001) were IL-1B, IL-6, and TNF. Biological pathways regulated in module 4 included acute phase response signaling, coagulation system, and complement system (p < 0.001) (Figure 3).
Conclusion: We identified novel DRP in JDM involved in metabolic dysregulation (CAPN3 and IDO1), IFN signaling (CXCL10) and muscle structure and myogenesis. WCGNA identified a protein module associated with JDM skin and global disease activity enriched for proteins involved in acute phase response signaling and complement system. Further study of candidate proteins may assist in development of personalized care for JDM patients.
 Heat map of differentially regulated proteins in treatment naïve JDM patients relative to active JDM on treatment and CTL (FDR < 0.10). Each column represents an individual patient sample while each row represents a differentially expressed protein in treatment naïve JDM relative to CTL and active disease on treatment. Included DRP were those with greatest fold change from each sub-panel from the Olink® Explore 3072 panel. Protein expression values are depicted using the color scale shown with purple to yellow indicating increasing expression.
Heat map of differentially regulated proteins in treatment naïve JDM patients relative to active JDM on treatment and CTL (FDR < 0.10). Each column represents an individual patient sample while each row represents a differentially expressed protein in treatment naïve JDM relative to CTL and active disease on treatment. Included DRP were those with greatest fold change from each sub-panel from the Olink® Explore 3072 panel. Protein expression values are depicted using the color scale shown with purple to yellow indicating increasing expression. .jpg) Correlation of candidate protein biomarkers with PGA and CDASI (p-value < 0.001)
Correlation of candidate protein biomarkers with PGA and CDASI (p-value < 0.001).jpg) Predominant regulated pathways from proteins included in module 4 identified using WGCNA. The dot size represents the number of proteins regulated in each pathway compared to the total number of proteins in the pathway (protein ratio). The dot color represents the -log(p-value).
Predominant regulated pathways from proteins included in module 4 identified using WGCNA. The dot size represents the number of proteins regulated in each pathway compared to the total number of proteins in the pathway (protein ratio). The dot color represents the -log(p-value). Disclosures: M. Kerski, None; C. Berthier, None; A. Tsoi, None; S. Vandenbergen, None; M. McClune, None; C. Powell, None; J. Kahlenberg, AstraZeneca, Bristol-Myers Squibb(BMS), Eli Lilly, GlaxoSmithKlein(GSK), Janssen, Merck/MSD, Gilead; J. Turnier, None.

