Back
Poster Session A
Systemic lupus erythematosus (SLE)
Session: (0317–0342) SLE – Diagnosis, Manifestations, and Outcomes Poster I: Diagnosis
0336: Machine Learning Approach for the Prediction of Lupus Nephritis Renal Flares Using Polygenic Risk Score and Electronic Health Record
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- YC
Yi-Ming Chen, MD
Taichung Veterans General Hospital, Taiwan
Taichung, Taiwan
Abstract Poster Presenter(s)
Yi-Ming Chen1, Tzu-Hung Hsiao1, Seng-Cho Chou2 and Chi-Wei Chung2, 1Taichung Veterans General Hospital, Taichung, Taiwan, 2Department of Information Management, National Taiwan University, Taipei, Taiwan
Background/Purpose: Lupus nephritis (LN) flares are associated with risks of renal failure and mortality in patients with systemic lupus erythematous (SLE). It is critical to stratify risks and provide individual care plans. Our study aimed to construct machine learning (ML) models for prediction of LN flares.
Methods: A total of 1,546 patients with were enrolled from the Taichung Veterans General Hospital. Genome-wide single nucleotide polymorphism (SNP) data were obtained using Taiwan Biobank version 2 array. Electronic health record (EHR), SNP and polygenic risk score (PRS) were combined to construct a proposed hybrid ML models using logistic regression, random forest, support vector machine, extreme gradient boosting (XGB) and light gradient boosting Machine (Figure 1). SHapley Additive exPlanation (SHAP) values were calculated to determine the contribution of each feature.
Results: In total, we enrolled 1,564 patients with SLE; 448 patients experienced LN flare within 5 years. Among 686,354 SNPs, 375 SNPs with p-value of genome-wide significant threshold < 1 × 10-3 were used to compute the PRS (Figure 2A). Figure 2B & C illustrated the comparisons of area under the receiver operating characteristic (AUROC) and area under the precision-recall curve (AUPRC) of models using SNP alone (SNP012), SNP PRS and a modified summation of PRS with weighted by p-value (SNP PRS+PRSw). In addition, we discovered that the model performance using both EHR and genomic features (SNP PRS+PRSw) exhibited the highest AUROC of 0.9512 and AUPRC of 0.8902 in validation set (Figure 2D & E). Our results showed the proposed hybrid framework combined the classifier model of XGB could achieve the highest result with AUPRC of 0.9021 in unseen testing set. SHAP summary plot depicted top 20 features predicting LN flare (Figure 3).
Conclusion: We established robust ML methods for prediction of LN flares using a novel hybrid combination of SNP, PRS and EHR. Future studies are required to confirm our findings and unpin the mechanistic explanation.
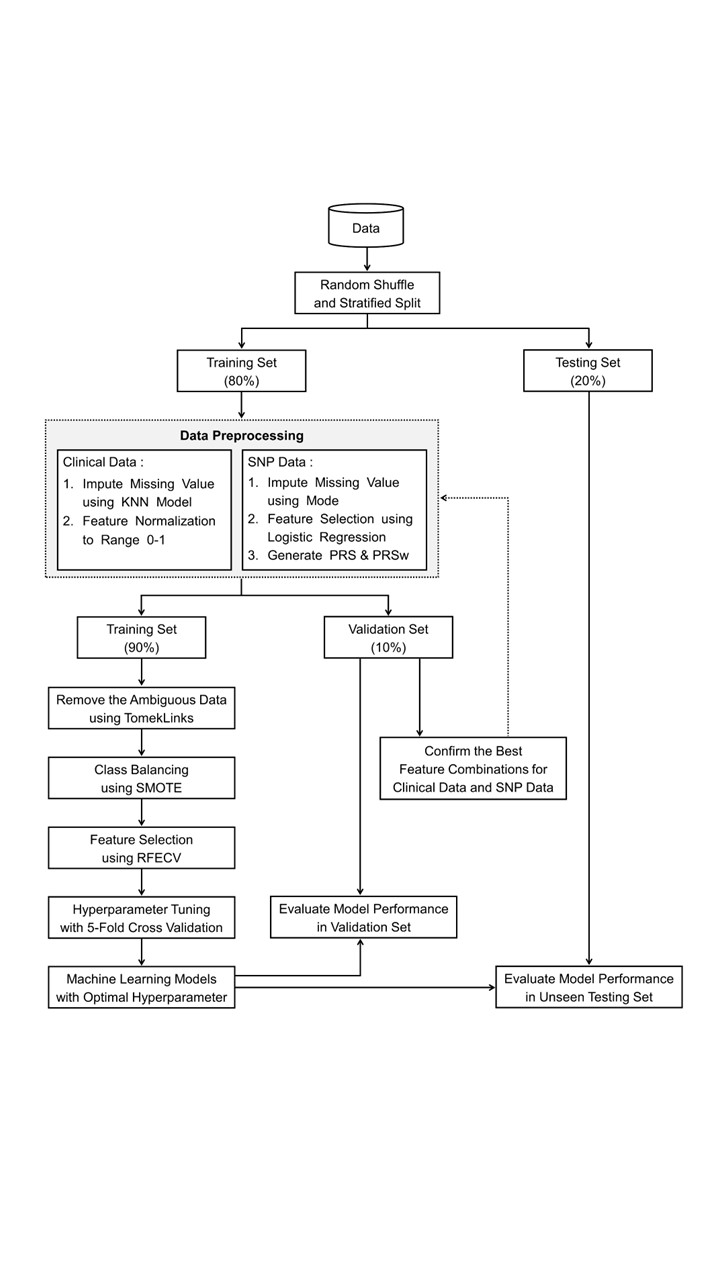 Figure 1. Schematic diagram of the proposed hybrid approach
Figure 1. Schematic diagram of the proposed hybrid approach
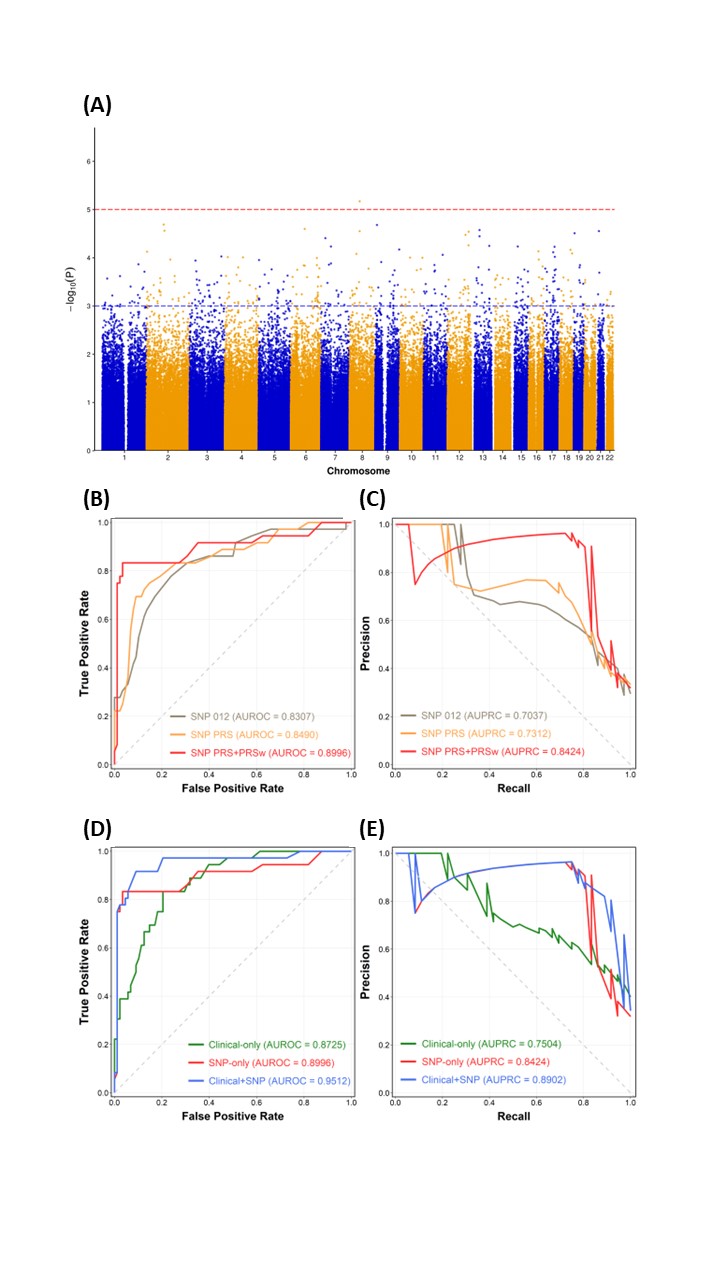 Figure 2. (A) Manhattan plot of GWAS between LN and non-LN patients; (B) ROC curve and (C) PR curve of comparisons in model performance among three SNP feature combinations; (D) ROC curve and (E) PR curve of comparisons of model performance among three feature combinations using clinical and SNP data.
Figure 2. (A) Manhattan plot of GWAS between LN and non-LN patients; (B) ROC curve and (C) PR curve of comparisons in model performance among three SNP feature combinations; (D) ROC curve and (E) PR curve of comparisons of model performance among three feature combinations using clinical and SNP data.
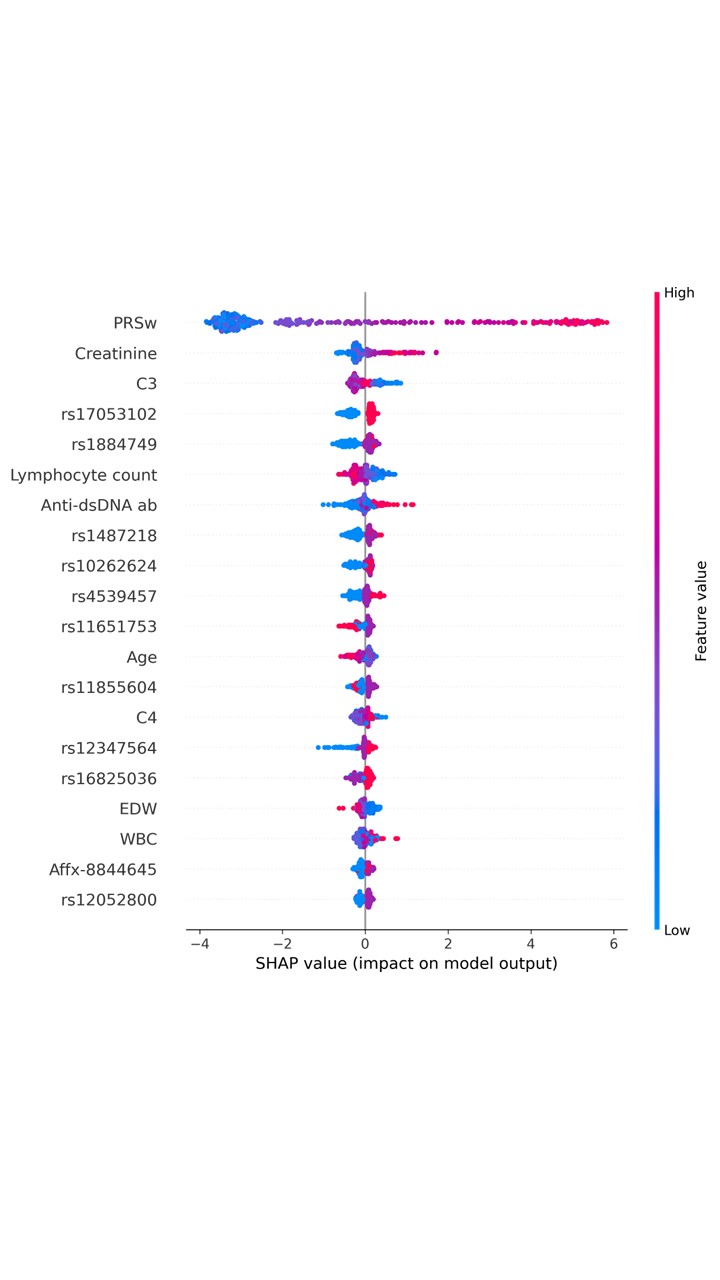 Figure 3. SHAP summary plot of top 20 features of the proposed hybrid framework with XGB model. PRSw: modified PRS weighted by the p-value; Anti-dsDNA ab: anti-dsDNA antibody; EDW: erythrocyte distribution width; WBC: white blood cell.
Figure 3. SHAP summary plot of top 20 features of the proposed hybrid framework with XGB model. PRSw: modified PRS weighted by the p-value; Anti-dsDNA ab: anti-dsDNA antibody; EDW: erythrocyte distribution width; WBC: white blood cell.
Disclosures: Y. Chen, None; T. Hsiao, None; S. Chou, None; C. Chung, None.
Background/Purpose: Lupus nephritis (LN) flares are associated with risks of renal failure and mortality in patients with systemic lupus erythematous (SLE). It is critical to stratify risks and provide individual care plans. Our study aimed to construct machine learning (ML) models for prediction of LN flares.
Methods: A total of 1,546 patients with were enrolled from the Taichung Veterans General Hospital. Genome-wide single nucleotide polymorphism (SNP) data were obtained using Taiwan Biobank version 2 array. Electronic health record (EHR), SNP and polygenic risk score (PRS) were combined to construct a proposed hybrid ML models using logistic regression, random forest, support vector machine, extreme gradient boosting (XGB) and light gradient boosting Machine (Figure 1). SHapley Additive exPlanation (SHAP) values were calculated to determine the contribution of each feature.
Results: In total, we enrolled 1,564 patients with SLE; 448 patients experienced LN flare within 5 years. Among 686,354 SNPs, 375 SNPs with p-value of genome-wide significant threshold < 1 × 10-3 were used to compute the PRS (Figure 2A). Figure 2B & C illustrated the comparisons of area under the receiver operating characteristic (AUROC) and area under the precision-recall curve (AUPRC) of models using SNP alone (SNP012), SNP PRS and a modified summation of PRS with weighted by p-value (SNP PRS+PRSw). In addition, we discovered that the model performance using both EHR and genomic features (SNP PRS+PRSw) exhibited the highest AUROC of 0.9512 and AUPRC of 0.8902 in validation set (Figure 2D & E). Our results showed the proposed hybrid framework combined the classifier model of XGB could achieve the highest result with AUPRC of 0.9021 in unseen testing set. SHAP summary plot depicted top 20 features predicting LN flare (Figure 3).
Conclusion: We established robust ML methods for prediction of LN flares using a novel hybrid combination of SNP, PRS and EHR. Future studies are required to confirm our findings and unpin the mechanistic explanation.
 Figure 1. Schematic diagram of the proposed hybrid approach
Figure 1. Schematic diagram of the proposed hybrid approach Figure 2. (A) Manhattan plot of GWAS between LN and non-LN patients; (B) ROC curve and (C) PR curve of comparisons in model performance among three SNP feature combinations; (D) ROC curve and (E) PR curve of comparisons of model performance among three feature combinations using clinical and SNP data.
Figure 2. (A) Manhattan plot of GWAS between LN and non-LN patients; (B) ROC curve and (C) PR curve of comparisons in model performance among three SNP feature combinations; (D) ROC curve and (E) PR curve of comparisons of model performance among three feature combinations using clinical and SNP data. Figure 3. SHAP summary plot of top 20 features of the proposed hybrid framework with XGB model. PRSw: modified PRS weighted by the p-value; Anti-dsDNA ab: anti-dsDNA antibody; EDW: erythrocyte distribution width; WBC: white blood cell.
Figure 3. SHAP summary plot of top 20 features of the proposed hybrid framework with XGB model. PRSw: modified PRS weighted by the p-value; Anti-dsDNA ab: anti-dsDNA antibody; EDW: erythrocyte distribution width; WBC: white blood cell.Disclosures: Y. Chen, None; T. Hsiao, None; S. Chou, None; C. Chung, None.

