Back
Poster Session A
Systemic lupus erythematosus (SLE)
Session: (0317–0342) SLE – Diagnosis, Manifestations, and Outcomes Poster I: Diagnosis
0334: Performance of Diagnosis Codes for Identification of Prevalent Lupus Nephritis Patients from Electronic Health Records
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- ZI
Zara Izadi, PhD, PharmD
University of California San Francisco
San Francisco, CA, United States
Abstract Poster Presenter(s)
Zara Izadi1, Alfredo Aguirre2, Christine Anastasiou2, Julia Kay3, Gabriela Schmajuk4 and Jinoos Yazdany5, 1University of California San Francisco, San Francisco, CA, 2University of California, San Francisco, San Francisco, CA, 3UCSF, San Francisco, CA, 4UCSF / SFVA, San Francisco, CA, 5UCSF, San Rafael, CA
Background/Purpose: Real-world data could provide valuable information in the study of lupus nephritis (LN), if these cases could be reliably identified. A lack of ability to perform chart reviews has prohibited a detailed understanding of under-coding of LN diagnoses in EHRs. In this study we aimed to determine the current performance of ICD9/10 codes for identifying prevalent LN, using a manual chart review of the EHR as gold standard.
Methods: We used structured data and notes from EHRs of two large health systems in the San Francisco Bay Area and included patients with ≥1 ICD9/10 codes for SLE from June 2012 to Jan 2022. Prevalent LN was defined as any documentation of LN diagnosis in EHR notes which included active LN and LN in remission. We used a combination of text extraction using regular expressions (of "lupus nephritis" and related terms) and a manual structured chart review on 100 patients from each health system to identify LN cases (Figure 1). The chart review was performed by two rheumatologists with an independent rheumatologist adjudicating any disagreements. For each health system, in order to enrich chart review with sufficient LN cases, we included a random sample of 50 patients and a random sample of 50 "possible" LN patients defined as either 1) any renal ICD codes, or 2) ≥1 urine protein to creatinine ratio ≥0.5 mg/mg. Patients were categorized into 'no LN', 'definite LN' (biopsy report and LN Class III, IV, or V verified), 'potential LN' (no biopsy report but physician diagnosed LN), and 'diagnostic uncertainty' (physician states LN is possible). For each health system separately, we determined the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of ≥1 ICD9/10 codes and ≥2 ICD9/10 codes ≥30 days apart, for identifying patients with a "strict" (definite LN) or an "inclusive" (definite LN, potential LN, or diagnostic uncertainty) definition of LN.
Results: The random samples for manual chart review were selected from a total of 4,522 patients from both health systems (Table 1). A majority of chart-reviewed cases had definite LN and were identified with the ICD10 codes M32.14 or M32.15, or ICD9 code 710.0 in combination with ICD9 codes 583.81, 581.81, or 583.89. The total number of chart-reviewed cases with a strict and inclusive definition of LN was 34 and 41 (out of 100) from health system A, and 40 and 46 (out of 100) from health system B. The sensitivity of ≥1 ICD9/10 codes based on a strict and inclusive definition of LN was 55.9% and 51.2% at health system A, and 72.5% and 65.2% at health system B. The specificity, PPV and NPV of ≥1 ICD9/10 codes based on a strict definition of LN was 93.9%, 82.6%, and 80.5% at health system A and 91.6%, 85.3%, and 83.3% at health system B. The diagnostic criterion of ≥1 ICD9/10 codes used with the strict definition of LN yielded the highest accuracy in both health systems (Table 2).
Conclusion: Despite good specificity and PPV, the sensitivity of diagnosis codes for identification of LN was low, ranging from 42.5% to 72.5%. Performance of diagnosis codes varied by diagnostic criteria, definition of LN, and across health systems. Further work is needed to address under-coding of LN diagnoses in order to identify incident and prevalent cases of LN.
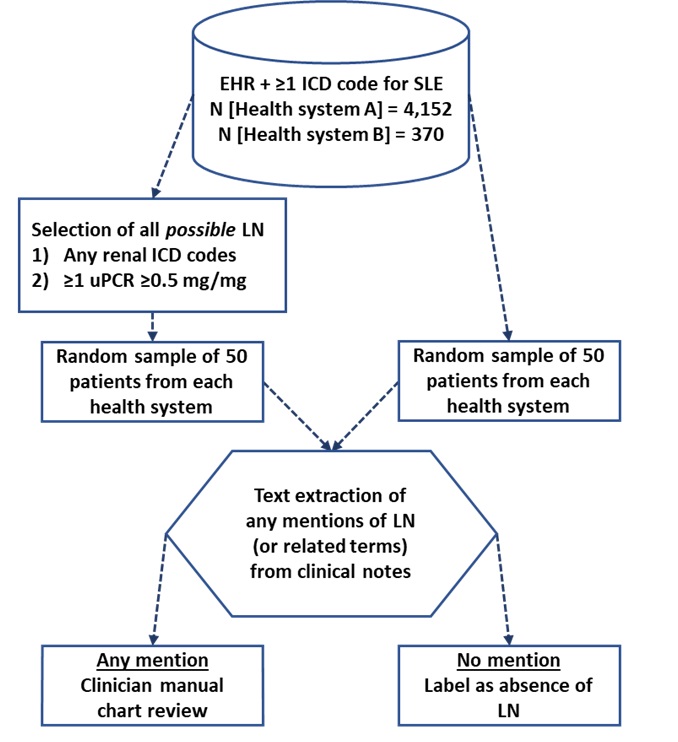 Overview of methodology for the gold-standard set development.
Overview of methodology for the gold-standard set development.
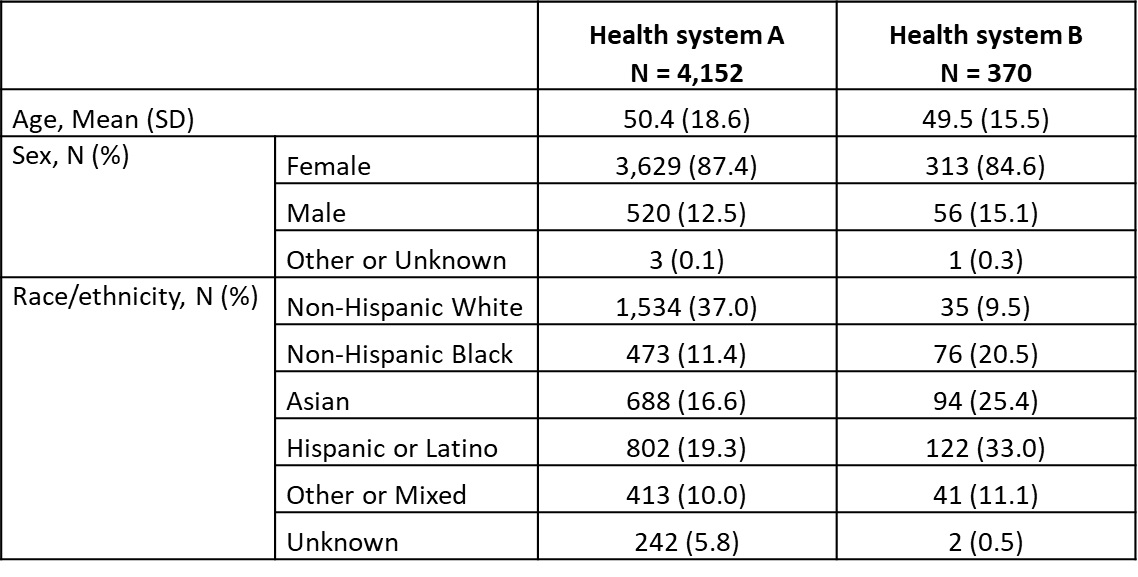 Characteristics of the underlying population.
Characteristics of the underlying population.
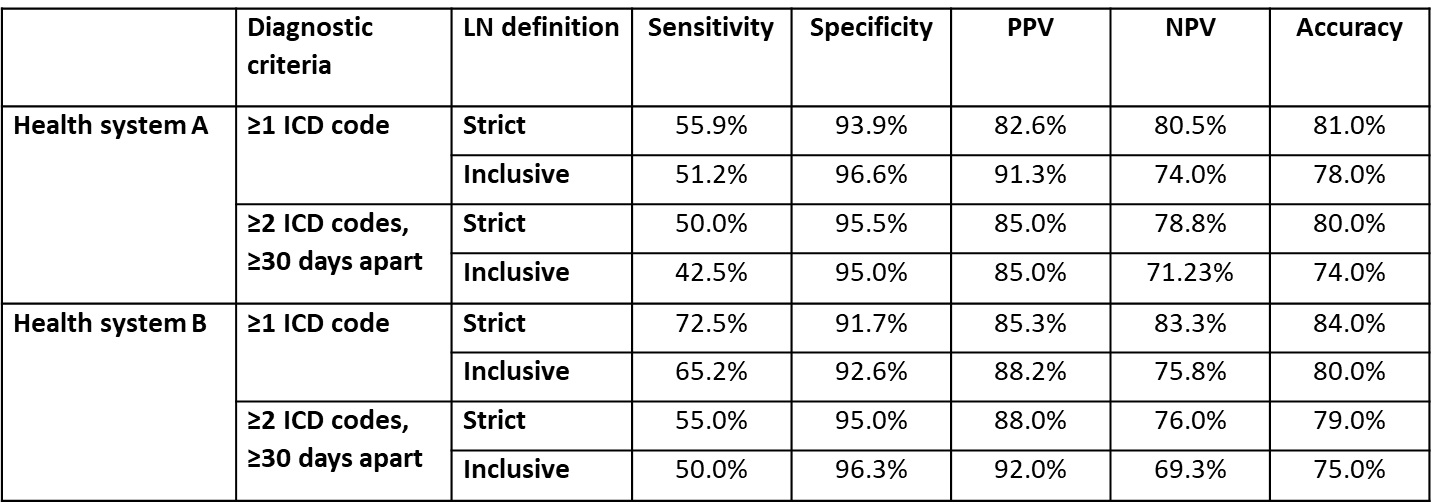 Performance of diagnosis codes for identifying prevalent LN patients. Strict: Definite LN only; Inclusive: definite LN, potential LN, or diagnostic uncertainty; PPV: positive predictive value; NPV: negative predictive value.
Performance of diagnosis codes for identifying prevalent LN patients. Strict: Definite LN only; Inclusive: definite LN, potential LN, or diagnostic uncertainty; PPV: positive predictive value; NPV: negative predictive value.
Disclosures: Z. Izadi, None; A. Aguirre, None; C. Anastasiou, None; J. Kay, None; G. Schmajuk, None; J. Yazdany, AstraZeneca, AstraZeneca, Bristol-Myers Squibb(BMS), Gilead, Pfizer, Aurinia.
Background/Purpose: Real-world data could provide valuable information in the study of lupus nephritis (LN), if these cases could be reliably identified. A lack of ability to perform chart reviews has prohibited a detailed understanding of under-coding of LN diagnoses in EHRs. In this study we aimed to determine the current performance of ICD9/10 codes for identifying prevalent LN, using a manual chart review of the EHR as gold standard.
Methods: We used structured data and notes from EHRs of two large health systems in the San Francisco Bay Area and included patients with ≥1 ICD9/10 codes for SLE from June 2012 to Jan 2022. Prevalent LN was defined as any documentation of LN diagnosis in EHR notes which included active LN and LN in remission. We used a combination of text extraction using regular expressions (of "lupus nephritis" and related terms) and a manual structured chart review on 100 patients from each health system to identify LN cases (Figure 1). The chart review was performed by two rheumatologists with an independent rheumatologist adjudicating any disagreements. For each health system, in order to enrich chart review with sufficient LN cases, we included a random sample of 50 patients and a random sample of 50 "possible" LN patients defined as either 1) any renal ICD codes, or 2) ≥1 urine protein to creatinine ratio ≥0.5 mg/mg. Patients were categorized into 'no LN', 'definite LN' (biopsy report and LN Class III, IV, or V verified), 'potential LN' (no biopsy report but physician diagnosed LN), and 'diagnostic uncertainty' (physician states LN is possible). For each health system separately, we determined the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of ≥1 ICD9/10 codes and ≥2 ICD9/10 codes ≥30 days apart, for identifying patients with a "strict" (definite LN) or an "inclusive" (definite LN, potential LN, or diagnostic uncertainty) definition of LN.
Results: The random samples for manual chart review were selected from a total of 4,522 patients from both health systems (Table 1). A majority of chart-reviewed cases had definite LN and were identified with the ICD10 codes M32.14 or M32.15, or ICD9 code 710.0 in combination with ICD9 codes 583.81, 581.81, or 583.89. The total number of chart-reviewed cases with a strict and inclusive definition of LN was 34 and 41 (out of 100) from health system A, and 40 and 46 (out of 100) from health system B. The sensitivity of ≥1 ICD9/10 codes based on a strict and inclusive definition of LN was 55.9% and 51.2% at health system A, and 72.5% and 65.2% at health system B. The specificity, PPV and NPV of ≥1 ICD9/10 codes based on a strict definition of LN was 93.9%, 82.6%, and 80.5% at health system A and 91.6%, 85.3%, and 83.3% at health system B. The diagnostic criterion of ≥1 ICD9/10 codes used with the strict definition of LN yielded the highest accuracy in both health systems (Table 2).
Conclusion: Despite good specificity and PPV, the sensitivity of diagnosis codes for identification of LN was low, ranging from 42.5% to 72.5%. Performance of diagnosis codes varied by diagnostic criteria, definition of LN, and across health systems. Further work is needed to address under-coding of LN diagnoses in order to identify incident and prevalent cases of LN.
 Overview of methodology for the gold-standard set development.
Overview of methodology for the gold-standard set development.  Characteristics of the underlying population.
Characteristics of the underlying population.  Performance of diagnosis codes for identifying prevalent LN patients. Strict: Definite LN only; Inclusive: definite LN, potential LN, or diagnostic uncertainty; PPV: positive predictive value; NPV: negative predictive value.
Performance of diagnosis codes for identifying prevalent LN patients. Strict: Definite LN only; Inclusive: definite LN, potential LN, or diagnostic uncertainty; PPV: positive predictive value; NPV: negative predictive value. Disclosures: Z. Izadi, None; A. Aguirre, None; C. Anastasiou, None; J. Kay, None; G. Schmajuk, None; J. Yazdany, AstraZeneca, AstraZeneca, Bristol-Myers Squibb(BMS), Gilead, Pfizer, Aurinia.

