Back
Poster Session A
Systemic lupus erythematosus (SLE)
Session: (0317–0342) SLE – Diagnosis, Manifestations, and Outcomes Poster I: Diagnosis
0329: Scoring Personalized Molecular Portraits Identify Systemic Lupus Erythematosus Subtypes and Predict Individualized Drug Responses, Symptomatology and Disease Progression
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- DT
Daniel Toro-Domínguez, Sr, PhD
GENYO
Granada, Spain
Abstract Poster Presenter(s)
Daniel Toro-Domínguez1, Manuel Martinez-Bueno1, Raúl López-Domínguez2, Jordi Martorell-Marugán1, Elena Carnero-Montoro1, Guillermo Barturen1, Daniel W. Goldman3, Michelle Petri3, Pedro Carmona-Sáez2 and Marta Alarcon-Riquelme1, 1Center for Genomics and Oncological Research (GENYO), Granada, Spain, 2University of Granada, Granada, Spain, 3Johns Hopkins University School of Medicine, Division of Rheumatology, Baltimore, MD
Background/Purpose: Systemic Lupus Erythematosus is a complex autoimmune disease that leads to important worsening of quality of life and mortality. Flares appear unpredictably during the disease course and therapies used are often only partially effective. These challenges are mainly due to the molecular heterogeneity of the disease, such that personalized medicine offers major promise. With this work we intended to advance in that direction by developing MyPROSLE, an omic-based workflow for measuring the molecular portrait of individual patients to support clinicians in their therapeutic decisions.
Methods: Immunological gene-modules were used to represent the transcriptome of the patients. A dysregulation score for each gene-module was calculated at the patient level based on averaged z-scores (M-scores). Almost 6100 lupus and 750 healthy samples were used to analyze the association between dysregulation scores, clinical manifestations, prognosis, flare and remission events and drug response to Tabalumab. Machine learning-based classification models were built to predict around 100 different clinical parameters based on personalized dysregulation scores. The system was called MyPROSLE.
Results: MyPROSLE allows to molecularly summarize patients in 206 gene-modules, clustered into 9 main lupus signatures, the combination of which revealed highly differentiated pathological mechanisms. We show that dysregulation of certain gene-modules is strongly associated with specific clinical manifestations, the occurrence of relapses or the potential presence of long-term remission and drug response. Therefore, MyPROSLE could be used to accurately predict these clinical outcomes, example, we can predict severe nephritis (AUC = 0.98) without the need for biopsy.
Conclusion: MyPROSLE (available at https://myprosle.genyo.es) allows molecular characterization of individual Lupus patients and it extracts key molecular information to support more precise therapeutic decisions. Figure 1 shows a summary of the main steps performed on the web tool.
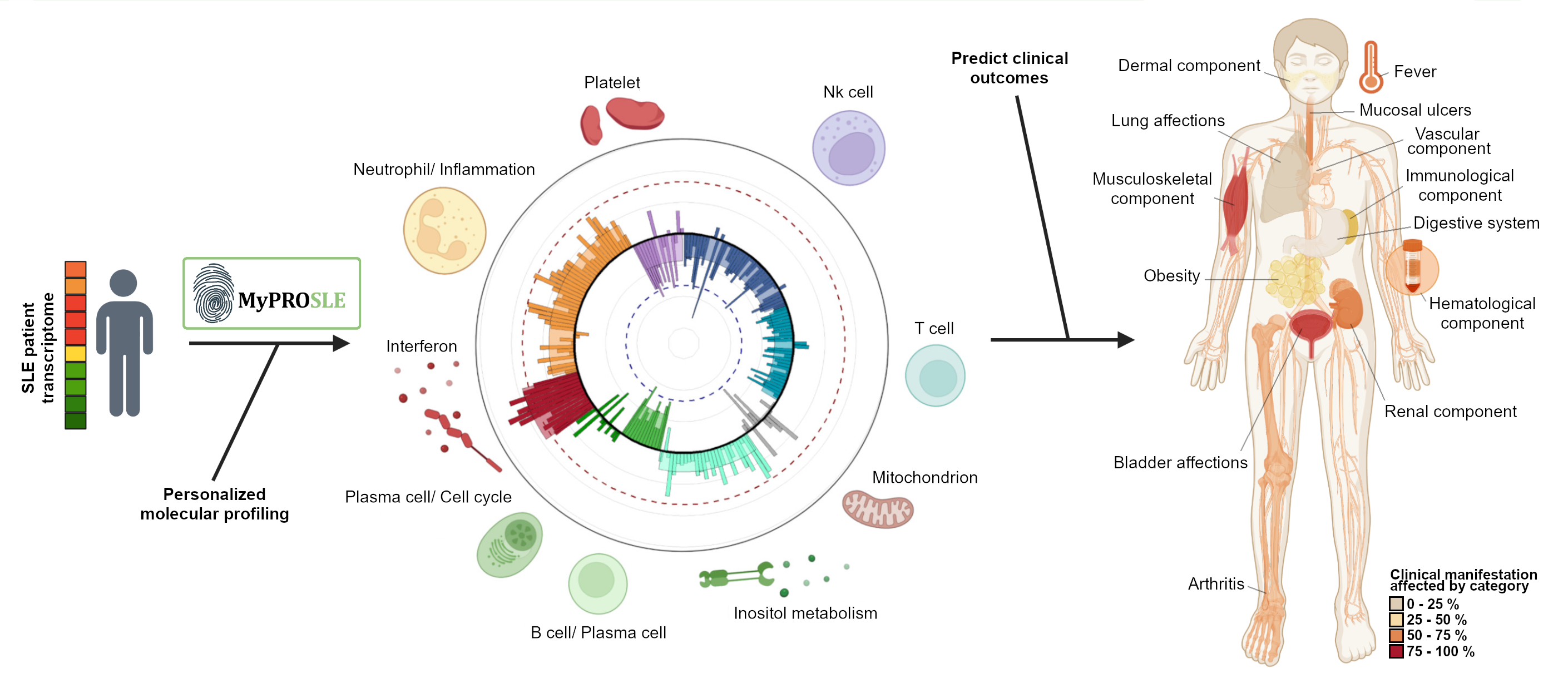 Figure 1. Web tool output example. The figure shows a summary of the web output for each of the two main steps, the personalized molecular profiling of the patients by calculating the M-scores and the clinical outcomes prediction.
Figure 1. Web tool output example. The figure shows a summary of the web output for each of the two main steps, the personalized molecular profiling of the patients by calculating the M-scores and the clinical outcomes prediction.
Disclosures: D. Toro-Domínguez, None; M. Martinez-Bueno, None; R. López-Domínguez, None; J. Martorell-Marugán, None; E. Carnero-Montoro, None; G. Barturen, None; D. Goldman, None; M. Petri, Exagen, AstraZeneca, Alexion, Amgen, AnaptysBio, Argenx, Aurinia, Biogen, Caribou Biosciences, CVS Health, EMD Serono, Eli Lilly, Emergent Biosolutions, GlaxoSmithKline (GSK), IQVIA, Janssen, Kira Pharmaceuticals, MedShr, Sanofi, SinoMab, Thermofisher, BPR Scientific Advisory Committee; P. Carmona-Sáez, None; M. Alarcon-Riquelme, None.
Background/Purpose: Systemic Lupus Erythematosus is a complex autoimmune disease that leads to important worsening of quality of life and mortality. Flares appear unpredictably during the disease course and therapies used are often only partially effective. These challenges are mainly due to the molecular heterogeneity of the disease, such that personalized medicine offers major promise. With this work we intended to advance in that direction by developing MyPROSLE, an omic-based workflow for measuring the molecular portrait of individual patients to support clinicians in their therapeutic decisions.
Methods: Immunological gene-modules were used to represent the transcriptome of the patients. A dysregulation score for each gene-module was calculated at the patient level based on averaged z-scores (M-scores). Almost 6100 lupus and 750 healthy samples were used to analyze the association between dysregulation scores, clinical manifestations, prognosis, flare and remission events and drug response to Tabalumab. Machine learning-based classification models were built to predict around 100 different clinical parameters based on personalized dysregulation scores. The system was called MyPROSLE.
Results: MyPROSLE allows to molecularly summarize patients in 206 gene-modules, clustered into 9 main lupus signatures, the combination of which revealed highly differentiated pathological mechanisms. We show that dysregulation of certain gene-modules is strongly associated with specific clinical manifestations, the occurrence of relapses or the potential presence of long-term remission and drug response. Therefore, MyPROSLE could be used to accurately predict these clinical outcomes, example, we can predict severe nephritis (AUC = 0.98) without the need for biopsy.
Conclusion: MyPROSLE (available at https://myprosle.genyo.es) allows molecular characterization of individual Lupus patients and it extracts key molecular information to support more precise therapeutic decisions. Figure 1 shows a summary of the main steps performed on the web tool.
 Figure 1. Web tool output example. The figure shows a summary of the web output for each of the two main steps, the personalized molecular profiling of the patients by calculating the M-scores and the clinical outcomes prediction.
Figure 1. Web tool output example. The figure shows a summary of the web output for each of the two main steps, the personalized molecular profiling of the patients by calculating the M-scores and the clinical outcomes prediction.Disclosures: D. Toro-Domínguez, None; M. Martinez-Bueno, None; R. López-Domínguez, None; J. Martorell-Marugán, None; E. Carnero-Montoro, None; G. Barturen, None; D. Goldman, None; M. Petri, Exagen, AstraZeneca, Alexion, Amgen, AnaptysBio, Argenx, Aurinia, Biogen, Caribou Biosciences, CVS Health, EMD Serono, Eli Lilly, Emergent Biosolutions, GlaxoSmithKline (GSK), IQVIA, Janssen, Kira Pharmaceuticals, MedShr, Sanofi, SinoMab, Thermofisher, BPR Scientific Advisory Committee; P. Carmona-Sáez, None; M. Alarcon-Riquelme, None.

