Back
Ignite Talk
Session: Ignite Session 1C
0658: Integrative Analysis of Cell-Specific Human Endogenous Retrovirus Expression and Host Gene Expression Identifies Associations with Clinical SLE Phenotypes
Saturday, November 12, 2022
1:45 PM – 1:50 PM Eastern Time
Location: South Philly Stage
- ZC
Zach Cutts, -None-
UCSF
San Francisco, CA, United StatesDisclosure: Disclosure information not submitted.
Ignite Speaker(s)
Zachary Cutts1, Sarah Patterson2, Lenka Maliskova1, Chun Ye2, Maria Dall'Era3, Jinoos Yazdany4, Lindsey Criswell5, Charles Langelier6, Marina Sirota7 and Cristina M Lanata8, 1University of California, San Francisco, San Francisco, 2University of California, San Francisco, San Francisco, CA, 3University of California, Division of Rheumatology, San Francisco, CA, 4UCSF, San Francisco, CA, 5National Human Genome Research Institute, NIH, Bethesda, MD, 6UCSF, San Francisco, 7UCSF, SF, 8NIH/NHGRI, Washington, DC
Background/Purpose: Human endogenous retroviruses (HERVs) and long interspersed nuclear elements (LINEs) make up 5-8% and 21% of the human genome, respectively. Their expression may contribute to production of type I interferon and the generation of autoantibodies. The objective of this study was to detect HERVs and LINEs in 4 cell-types in SLE patients and characterize their relationship to host gene expression and SLE phenotypes.
Methods: Peripheral blood mononuclear cells were isolated from 120 deeply phenotyped SLE participants. Cells were sorted utilizing magnetic beads (CD14+ monocytes, B cells, CD4+ T cells, and NK cells) and STEM cell technologies for a total of 480 samples. Libraries were sequenced on a HiSeq4000 PE150. Trimmed fastq files were aligned to GRCh38 release 104 using default settings with STAR to generate alignment files. Alignment files were converted to gene counts using featureCounts. Raw counts from Telescope were normalized using DESeq2 and summed per patient; patients were then separated into tertiles based on the summed counts for HERVs and LINEs. DESeq2 was used to perform differential gene expression analysis using gene counts from featureCounts, comparing the third to the first tertile. Gene set enrichment analysis was performed using genes with adjusted p values < 0.05, ranking genes by log2FoldChange, and running WebGestalt. For clinical outcomes, outliers were identified and dropped per cell type and differential expression analysis was run using raw counts from Telescope with DESeq2 per cell type, adjusting for race, lane, sex, and immunosuppressant use at the time of blood draw. Outcomes studied included disease activity (SLEDAI score), autoantibody production (dsDNA, RNP, Sm), ACR renal criteria and disease severity (as defined by clinical clusters previously described in the same SLE participants)1.
Results: A total of 26,768 HERVs/LINEs were detected across the 480 samples. These were mostly cell-specific (Fig1). High HERVs/LINEs expression correlated with host gene transcription in a cell specific manner. Differentially enriched pathways with retroviral load include olfactory signaling pathway, regulation of IFNA signaling, and interferon alpha/beta signaling in CD14 cells; DNA repair and host response of HIV factors in CD4 cells; activation of HOX genes and antimicrobial peptides in CD19 cells; and regulation of complement cascade, neutrophil degranulation and several metabolic pathways in NK cells (Fig 2). Significant associations between HERVs/LINEs expression and clinical outcomes are summarized in Table 1. We found that CD19 cells had the most robust associations with disease severity, SLEDAI score, history of renal disease, and autoantibody production (FDR p< 0.05). Other findings included high HERVs/LINEs expression in NK cells in patient with severe disease, and in CD4 cells in patients with dsDNA production (FDR p< 0.05).
Conclusion: HERVs/LINEs expression is associated with host gene expression in relevant immune pathways in patients with SLE, in a cell specific manner. Further, we found a strong association between HERVs/LINEs expression and clinical outcomes, particularly in CD19 cells, in SLE patients.
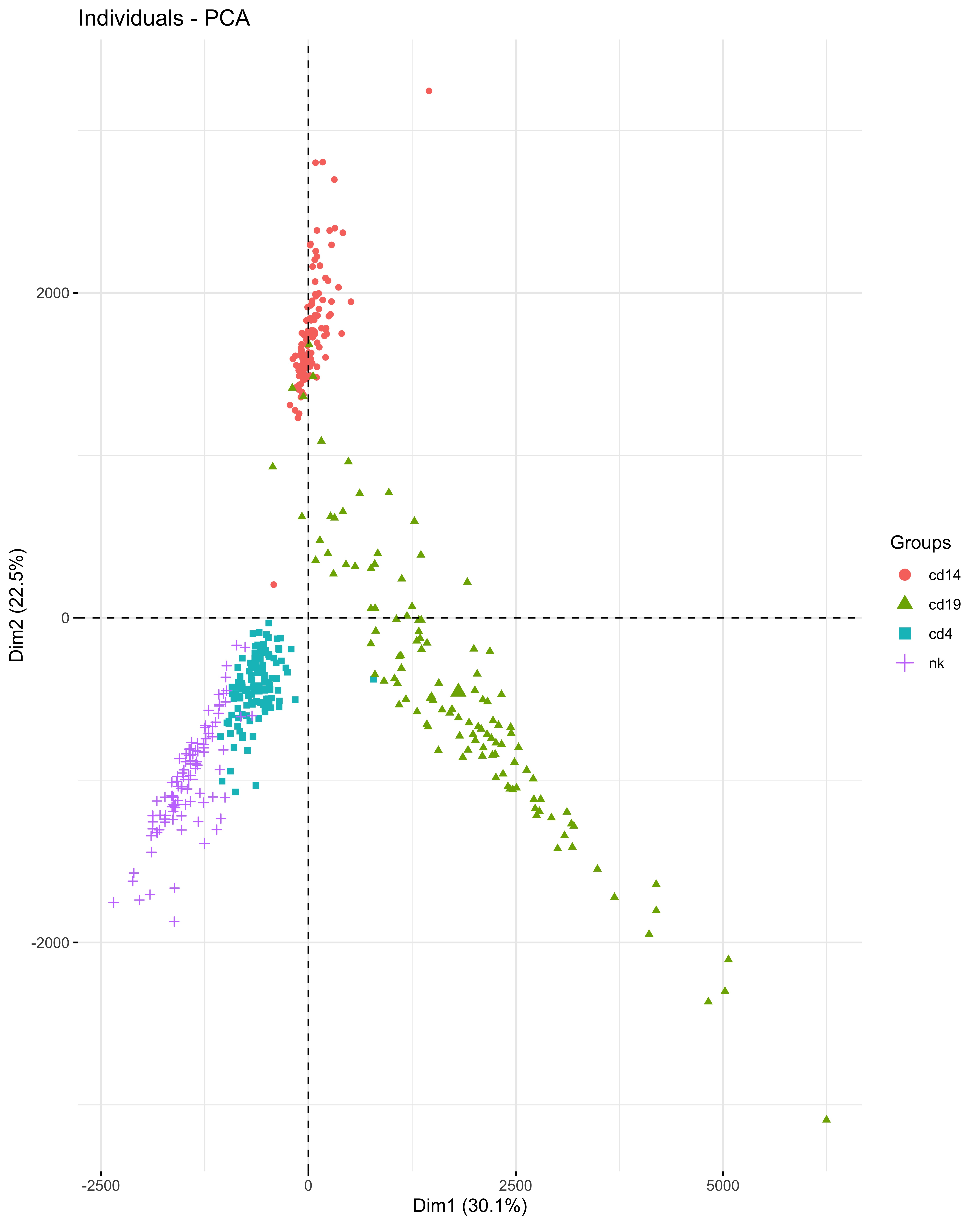 PCA plot of HERVs and LINEs expression in 4 cell types of 120 SLE individuals
PCA plot of HERVs and LINEs expression in 4 cell types of 120 SLE individuals
.jpg) Pathways of differentially expressed genes associated with high HERVs/LINEs expression per cell type in 120 SLE patients
Pathways of differentially expressed genes associated with high HERVs/LINEs expression per cell type in 120 SLE patients
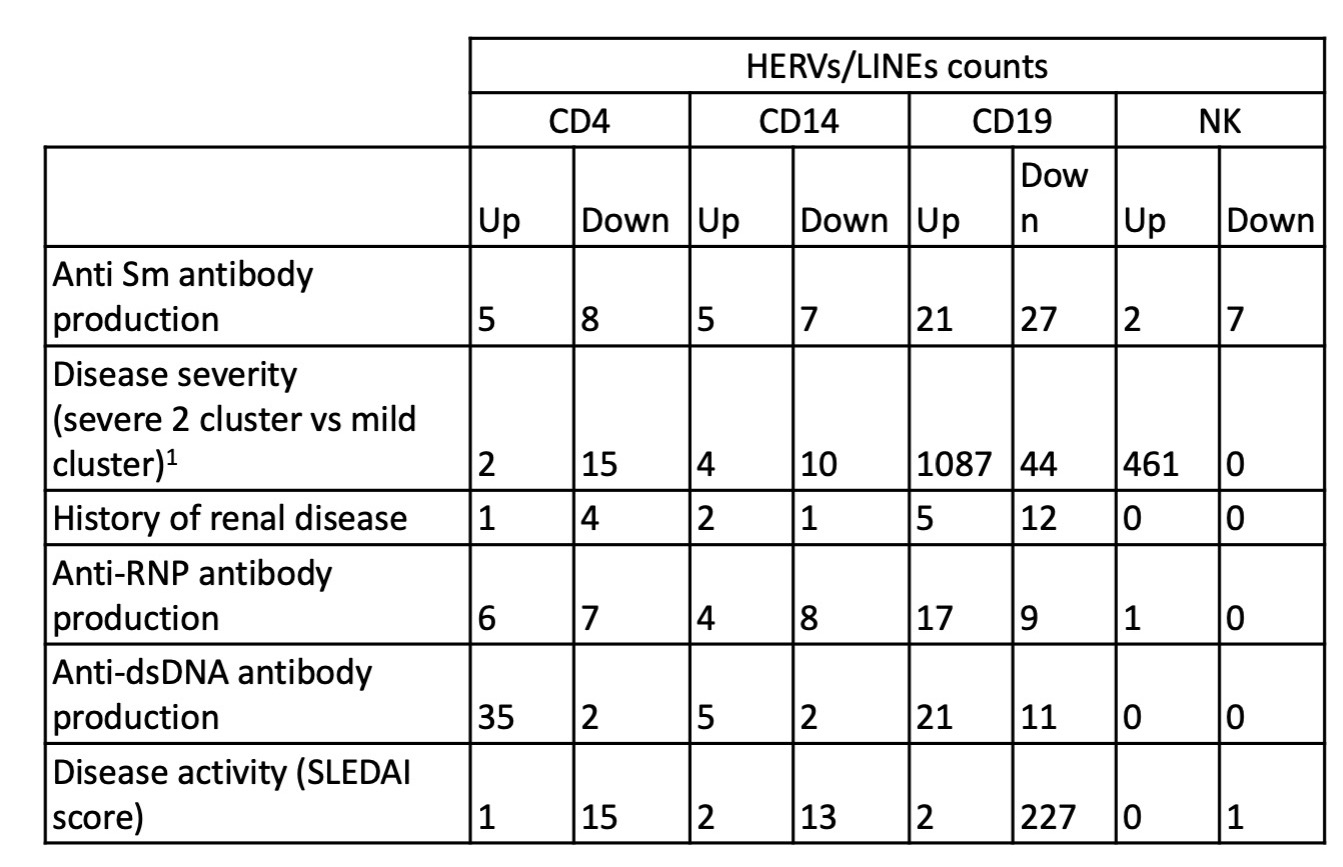 HERVs/LINEs expression associated with clinical outcomes, adjusted for sex, race and immunosuppressive medication use in 120 SLE participants
HERVs/LINEs expression associated with clinical outcomes, adjusted for sex, race and immunosuppressive medication use in 120 SLE participants
Disclosures: Z. Cutts, None; S. Patterson, Motto; L. Maliskova, None; C. Ye, None; M. Dall'Era, GlaxoSmithKline (GSK), AstraZeneca, Aurinia, BMS, Amgen; J. Yazdany, AstraZeneca, Gilead, Bristol-Myers Squibb(BMS), Aurinia, Astra Zeneca, Pfizer; L. Criswell, None; C. Langelier, None; M. Sirota, None; C. M Lanata, None.
Background/Purpose: Human endogenous retroviruses (HERVs) and long interspersed nuclear elements (LINEs) make up 5-8% and 21% of the human genome, respectively. Their expression may contribute to production of type I interferon and the generation of autoantibodies. The objective of this study was to detect HERVs and LINEs in 4 cell-types in SLE patients and characterize their relationship to host gene expression and SLE phenotypes.
Methods: Peripheral blood mononuclear cells were isolated from 120 deeply phenotyped SLE participants. Cells were sorted utilizing magnetic beads (CD14+ monocytes, B cells, CD4+ T cells, and NK cells) and STEM cell technologies for a total of 480 samples. Libraries were sequenced on a HiSeq4000 PE150. Trimmed fastq files were aligned to GRCh38 release 104 using default settings with STAR to generate alignment files. Alignment files were converted to gene counts using featureCounts. Raw counts from Telescope were normalized using DESeq2 and summed per patient; patients were then separated into tertiles based on the summed counts for HERVs and LINEs. DESeq2 was used to perform differential gene expression analysis using gene counts from featureCounts, comparing the third to the first tertile. Gene set enrichment analysis was performed using genes with adjusted p values < 0.05, ranking genes by log2FoldChange, and running WebGestalt. For clinical outcomes, outliers were identified and dropped per cell type and differential expression analysis was run using raw counts from Telescope with DESeq2 per cell type, adjusting for race, lane, sex, and immunosuppressant use at the time of blood draw. Outcomes studied included disease activity (SLEDAI score), autoantibody production (dsDNA, RNP, Sm), ACR renal criteria and disease severity (as defined by clinical clusters previously described in the same SLE participants)1.
Results: A total of 26,768 HERVs/LINEs were detected across the 480 samples. These were mostly cell-specific (Fig1). High HERVs/LINEs expression correlated with host gene transcription in a cell specific manner. Differentially enriched pathways with retroviral load include olfactory signaling pathway, regulation of IFNA signaling, and interferon alpha/beta signaling in CD14 cells; DNA repair and host response of HIV factors in CD4 cells; activation of HOX genes and antimicrobial peptides in CD19 cells; and regulation of complement cascade, neutrophil degranulation and several metabolic pathways in NK cells (Fig 2). Significant associations between HERVs/LINEs expression and clinical outcomes are summarized in Table 1. We found that CD19 cells had the most robust associations with disease severity, SLEDAI score, history of renal disease, and autoantibody production (FDR p< 0.05). Other findings included high HERVs/LINEs expression in NK cells in patient with severe disease, and in CD4 cells in patients with dsDNA production (FDR p< 0.05).
Conclusion: HERVs/LINEs expression is associated with host gene expression in relevant immune pathways in patients with SLE, in a cell specific manner. Further, we found a strong association between HERVs/LINEs expression and clinical outcomes, particularly in CD19 cells, in SLE patients.
 PCA plot of HERVs and LINEs expression in 4 cell types of 120 SLE individuals
PCA plot of HERVs and LINEs expression in 4 cell types of 120 SLE individuals.jpg) Pathways of differentially expressed genes associated with high HERVs/LINEs expression per cell type in 120 SLE patients
Pathways of differentially expressed genes associated with high HERVs/LINEs expression per cell type in 120 SLE patients HERVs/LINEs expression associated with clinical outcomes, adjusted for sex, race and immunosuppressive medication use in 120 SLE participants
HERVs/LINEs expression associated with clinical outcomes, adjusted for sex, race and immunosuppressive medication use in 120 SLE participantsDisclosures: Z. Cutts, None; S. Patterson, Motto; L. Maliskova, None; C. Ye, None; M. Dall'Era, GlaxoSmithKline (GSK), AstraZeneca, Aurinia, BMS, Amgen; J. Yazdany, AstraZeneca, Gilead, Bristol-Myers Squibb(BMS), Aurinia, Astra Zeneca, Pfizer; L. Criswell, None; C. Langelier, None; M. Sirota, None; C. M Lanata, None.

