Back
Abstract Session
Genetics, genomics and proteomics
Session: Abstracts: Genetics, Genomics and Proteomics (0562–0565)
0564: Identifying Shared Genetic Architecture Between RA and Other Conditions: A Phenome-Wide Association Study
Sunday, November 13, 2022
8:30 AM – 8:40 AM Eastern Time
Location: Room 120
- HZ
Harrison Zhang, BA
Brigham and Women's Hospital and Harvard Medical School
Boston, MA, United States
Presenting Author(s)
Harrison Zhang1, Gregory McDermott2, Thany Seyok2, Sicong Huang2, Kumar Dahal2, Sehi L'Yi1, Clara Lea-Bonzel1, Jacklyn Stratton2, Dana Weisenfeld2, Tianrun Cai2, Paul Monach3, Jing Cui2, Chuan Hong4, Tianxi Cai5 and Katherine Liao2, 1Harvard Medical School, Boston, MA, 2Brigham and Women's Hospital, Boston, MA, 3VA Boston Healthcare System, Boston, MA, 4Duke University, Durham, NC, 5Harvard TH Chan School of Public Health, Boston, MA
Background/Purpose: RA shares individual risk alleles with other autoimmune conditions such as type 1 diabetes and celiac disease. However, there are limited studies examining the association of all known RA risk alleles as a composite with other conditions. A broader screen to identify shared genetics across conditions can highlight potential shared pathways. The objective of this study was to identify conditions that share genetic architecture with RA by performing an RA genetic risk score (GRS) Phenome Wide Association Study (PheWAS) among non-RA patients.
Methods: We performed a PheWAS using data from the UK Biobank (UKB) with 319,946 patients. Using a previously published approach, we constructed 3 types of GRS: (1) a composite RA GRS combining HLA and non-HLA RA risk alleles; (2) HLA RA GRS; and (3) non-HLA RA GRS. Phenotypes were defined using published groupings of International Classification of Diseases (ICD) codes into clinically relevant codes, termed PheCodes. We tested the association between the composite RA GRS with n=1,491 phenotype groups using logistic regression adjusting for age, sex, self-reported race, and log-transformed number of hospital visits. Subjects with >=1 RA PheCode were excluded to decrease the potential for RA to confound the association. Statistical significance was defined using Bonferroni correction (0.05/1491). All findings were replicated at an academic center biobank, n=34,195. The accuracy of phenotype definitions was further validated with manual chart review using data from the institutional biobank, and positive predictive values (PPV) of PheCodes were calculated.
Results: We studied 318,946 non-RA UKB study subjects, mean age 70 years, 56% were female, 94% white. The UKB PheWAS identified 19 phenotypes significantly associated with the composite RA GRS, of which 13 (68%) were immune mediated conditions (Figure 1). Of these 19 phenotypes, 9 replicated in the academic biobank and had a PheCodes with PPV >0.80 from chart review (Table 1). Among these nine phenotypes, the composite RA GRS was associated with decreased risk of celiac disease (odds ratio [OR], 0.65; 95% CI, 0.61-0.70) and multiple sclerosis (MS, OR, 0.88; 95% CI, 0.84-0.92); and increased risk of PMR (OR, 1.32; 95% CI, 1.26-1.38) and granulomatosis with polyangiitis (GPA, OR, 1.30; 95% CI, 1.16-1.47). Known associations with type 1 diabetes, hypothyroidism, and diabetic retinopathy were also observed. The effects of HLA GRS and non-HLA GRS for most phenotypes shared the same direction of association, with the exception of celiac disease and MS (Figure 2). For celiac, the HLA RA GRS was associated with reduced risk and non-HLA was associated with increased risk.
Conclusion: In this population based PheWAS among non-RA patients, we identified novel associations between the RA GRS and reduced risk of MS, increased risk for PMR, and replicated a known association with increased risk for GPA. We additionally identified differential effects from HLA and non-HLA RA risk alleles on risk for celiac disease. Future directions include studies of the shared genetic variants between these conditions and whether they represent shared pathways early in the development of autoimmunity amenable for therapeutic targeting.
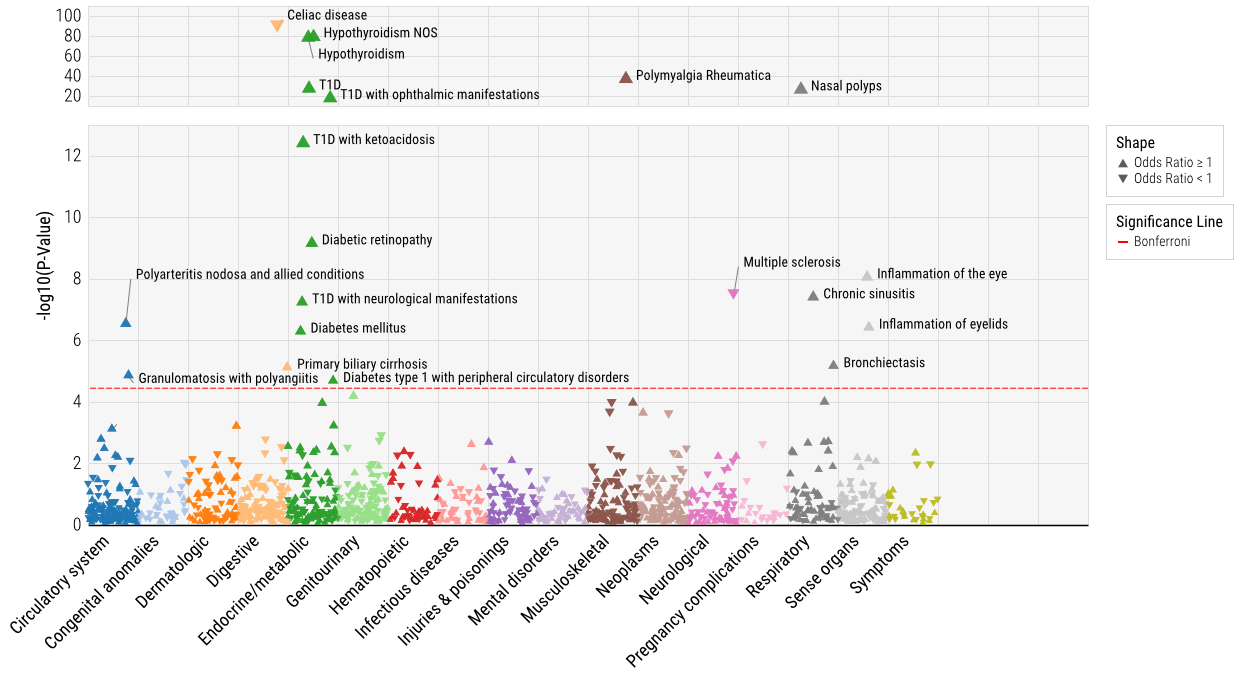 Figure 1. Phenome-Wide Association between the composite RA GRS with phenotypes in the U.K. Biobank.
Figure 1. Phenome-Wide Association between the composite RA GRS with phenotypes in the U.K. Biobank.
<img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1289595-2-ANY.jpg width=440 height=199.480519480519 border=0 style=border-style: none;>
<img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1289595-3-ANY.jpg width=440 height=317.826666666667 border=0 style=border-style: none;>
Disclosures: H. Zhang, None; G. McDermott, None; T. Seyok, None; S. Huang, None; K. Dahal, None; S. L'Yi, None; C. Lea-Bonzel, None; J. Stratton, None; D. Weisenfeld, None; T. Cai, None; P. Monach, Chemocentryx, Kiniksa, BMS/Celgene, Gilead; J. Cui, None; C. Hong, None; T. Cai, None; K. Liao, None.
Background/Purpose: RA shares individual risk alleles with other autoimmune conditions such as type 1 diabetes and celiac disease. However, there are limited studies examining the association of all known RA risk alleles as a composite with other conditions. A broader screen to identify shared genetics across conditions can highlight potential shared pathways. The objective of this study was to identify conditions that share genetic architecture with RA by performing an RA genetic risk score (GRS) Phenome Wide Association Study (PheWAS) among non-RA patients.
Methods: We performed a PheWAS using data from the UK Biobank (UKB) with 319,946 patients. Using a previously published approach, we constructed 3 types of GRS: (1) a composite RA GRS combining HLA and non-HLA RA risk alleles; (2) HLA RA GRS; and (3) non-HLA RA GRS. Phenotypes were defined using published groupings of International Classification of Diseases (ICD) codes into clinically relevant codes, termed PheCodes. We tested the association between the composite RA GRS with n=1,491 phenotype groups using logistic regression adjusting for age, sex, self-reported race, and log-transformed number of hospital visits. Subjects with >=1 RA PheCode were excluded to decrease the potential for RA to confound the association. Statistical significance was defined using Bonferroni correction (0.05/1491). All findings were replicated at an academic center biobank, n=34,195. The accuracy of phenotype definitions was further validated with manual chart review using data from the institutional biobank, and positive predictive values (PPV) of PheCodes were calculated.
Results: We studied 318,946 non-RA UKB study subjects, mean age 70 years, 56% were female, 94% white. The UKB PheWAS identified 19 phenotypes significantly associated with the composite RA GRS, of which 13 (68%) were immune mediated conditions (Figure 1). Of these 19 phenotypes, 9 replicated in the academic biobank and had a PheCodes with PPV >0.80 from chart review (Table 1). Among these nine phenotypes, the composite RA GRS was associated with decreased risk of celiac disease (odds ratio [OR], 0.65; 95% CI, 0.61-0.70) and multiple sclerosis (MS, OR, 0.88; 95% CI, 0.84-0.92); and increased risk of PMR (OR, 1.32; 95% CI, 1.26-1.38) and granulomatosis with polyangiitis (GPA, OR, 1.30; 95% CI, 1.16-1.47). Known associations with type 1 diabetes, hypothyroidism, and diabetic retinopathy were also observed. The effects of HLA GRS and non-HLA GRS for most phenotypes shared the same direction of association, with the exception of celiac disease and MS (Figure 2). For celiac, the HLA RA GRS was associated with reduced risk and non-HLA was associated with increased risk.
Conclusion: In this population based PheWAS among non-RA patients, we identified novel associations between the RA GRS and reduced risk of MS, increased risk for PMR, and replicated a known association with increased risk for GPA. We additionally identified differential effects from HLA and non-HLA RA risk alleles on risk for celiac disease. Future directions include studies of the shared genetic variants between these conditions and whether they represent shared pathways early in the development of autoimmunity amenable for therapeutic targeting.
 Figure 1. Phenome-Wide Association between the composite RA GRS with phenotypes in the U.K. Biobank.
Figure 1. Phenome-Wide Association between the composite RA GRS with phenotypes in the U.K. Biobank.<img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1289595-2-ANY.jpg width=440 height=199.480519480519 border=0 style=border-style: none;>
Table 1. Phenotypes associated with the composite RA GRS in the U.K. Biobank*.
*All nine phenotypes reported here were replicated in the academic biobank and had a PPV>0.8 from manual chart review.
*All nine phenotypes reported here were replicated in the academic biobank and had a PPV>0.8 from manual chart review.
<img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1289595-3-ANY.jpg width=440 height=317.826666666667 border=0 style=border-style: none;>
Figure 2. Association of RA HLA GRS and non-HLA GRS in the U.K. Biobank among nine phenotypes that were also validated in the academic biobank (and passed manual chart review with PPV>0.8).
Disclosures: H. Zhang, None; G. McDermott, None; T. Seyok, None; S. Huang, None; K. Dahal, None; S. L'Yi, None; C. Lea-Bonzel, None; J. Stratton, None; D. Weisenfeld, None; T. Cai, None; P. Monach, Chemocentryx, Kiniksa, BMS/Celgene, Gilead; J. Cui, None; C. Hong, None; T. Cai, None; K. Liao, None.

