Back
Poster Session D
Osteoarthritis (OA) and related disorders
Session: (1888–1923) Osteoarthritis – Clinical Poster
1898: Radiographic Osteoarthritis Progression Can Be Predicted via Pyrosequencing Analysis of Baseline Peripheral Blood
Monday, November 14, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- CD
Christopher Dunn, MSc
University of Oklahoma Health Sciences Center
Edmond, OK, United States
Abstract Poster Presenter(s)
Chris Dunn1, Cassandra Velasco2, Leoni Schlupp3, Emmaline Prinz3, Vladislav Izda4, Liubov Arbeeva5, Yvonne Golightly6, Amanda Nelson6 and Matlock Jeffries3, 1University of Oklahoma Health Sciences Center, Edmond, OK, 2University of Oklahoma Health Sciences Center, Oklahoma City, OK, 3Oklahoma Medical Research Foundation, Oklahoma City, OK, 4Oklahoma Medical Research Foundation, New York, NY, 5University of North Carolina Chapel Hill, Chapel Hill, NC, 6UNC School of Medicine, Chapel Hill, NC
Background/Purpose: Knee osteoarthritis (OA) is a heterogeneous disease characterized by a variety of clinical and molecular phenotypes. However, we do not yet have robust biomarkers to distinguish/predict these phenotypes. We previously published a pilot analysis of baseline peripheral blood cell DNA methylation patterns as biomarkers of future radiographic progression which was defined as an increase of ≥1 Kellgren-Lawrence Grade in the index between two visits. In the current study, we expand upon this methodology and apply it to the Johnston County Osteoarthritis Project (JoCoOA), targeting specific epigenetic regions for model development.
Methods: Buffy coat DNA was obtained from the JoCoOA (n=139) from a single time point prior to radiographic progression (5.6±1.1[mean±S.D.] years before progression). Demographic variables including age, race, gender and body mass index were also obtained and used in model development. DNA (500ng) was bisulfite treated, amplified and pyrosequenced. Pyrosequencing primers were designed to target regions around specific CpGs that were most useful for predicting progression in a previous array-based modeling study, allowing for quantification of regional DNA methylation changes that may be significant in predictive modeling. Data were divided into training (70%) and testing sets (30%) and generalized linear models (GLMs) were used for feature selection. Parsimonious models were applied to training and test set by including DNA methylation sites and covariates that had a p-value< 0.1 in initial model development.
Results: Baseline buffy coat DNA methylation patterns accurately predicted future radiographic progression of OA (AUC=0.80±0.010, accuracy=0.72±0.012 mean±standard error of mean [SEM], Table 1). Interestingly, the inclusion of demographic characteristics altered model performance. Specifically, inclusion of race increased the predictive capabilities of models (0.85±0.008 vs 0.80±0.010). Parsimonious models including only race and the top 3 CpG sites selected during full development had significantly higher predictive capabilities compared to models developed on all CpGs and demographic variables (accuracy p-value=0.00065).
Conclusion: Herein, we evaluated the predictive capability of peripheral blood-based DNA methylation models for radiographic OA progression in the JoCoOA. Future work should focus on evaluating differential DNA methylation of individual peripheral blood immune cell populations in individuals with knee OA.
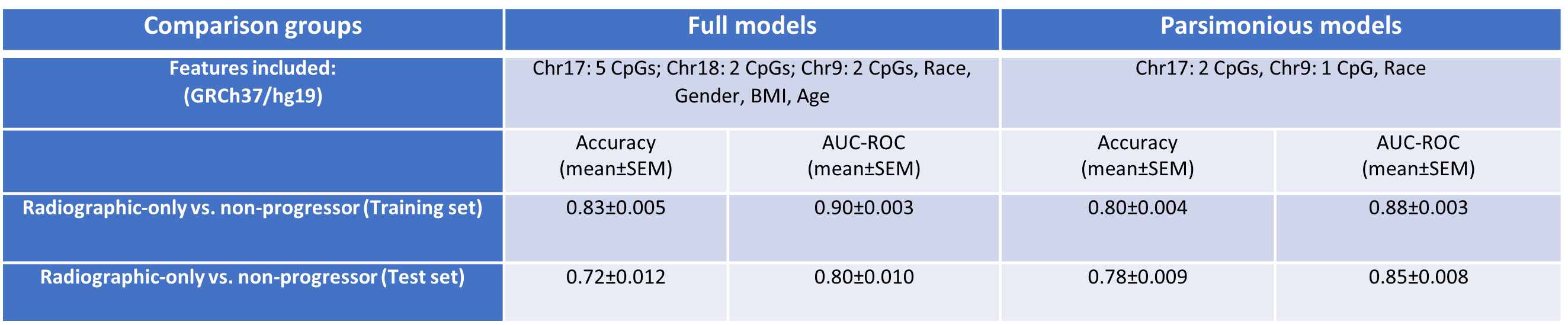 Model performance characteristics
Model performance characteristics
Disclosures: C. Dunn, None; C. Velasco, None; L. Schlupp, None; E. Prinz, None; V. Izda, None; L. Arbeeva, None; Y. Golightly, None; A. Nelson, None; M. Jeffries, None.
Background/Purpose: Knee osteoarthritis (OA) is a heterogeneous disease characterized by a variety of clinical and molecular phenotypes. However, we do not yet have robust biomarkers to distinguish/predict these phenotypes. We previously published a pilot analysis of baseline peripheral blood cell DNA methylation patterns as biomarkers of future radiographic progression which was defined as an increase of ≥1 Kellgren-Lawrence Grade in the index between two visits. In the current study, we expand upon this methodology and apply it to the Johnston County Osteoarthritis Project (JoCoOA), targeting specific epigenetic regions for model development.
Methods: Buffy coat DNA was obtained from the JoCoOA (n=139) from a single time point prior to radiographic progression (5.6±1.1[mean±S.D.] years before progression). Demographic variables including age, race, gender and body mass index were also obtained and used in model development. DNA (500ng) was bisulfite treated, amplified and pyrosequenced. Pyrosequencing primers were designed to target regions around specific CpGs that were most useful for predicting progression in a previous array-based modeling study, allowing for quantification of regional DNA methylation changes that may be significant in predictive modeling. Data were divided into training (70%) and testing sets (30%) and generalized linear models (GLMs) were used for feature selection. Parsimonious models were applied to training and test set by including DNA methylation sites and covariates that had a p-value< 0.1 in initial model development.
Results: Baseline buffy coat DNA methylation patterns accurately predicted future radiographic progression of OA (AUC=0.80±0.010, accuracy=0.72±0.012 mean±standard error of mean [SEM], Table 1). Interestingly, the inclusion of demographic characteristics altered model performance. Specifically, inclusion of race increased the predictive capabilities of models (0.85±0.008 vs 0.80±0.010). Parsimonious models including only race and the top 3 CpG sites selected during full development had significantly higher predictive capabilities compared to models developed on all CpGs and demographic variables (accuracy p-value=0.00065).
Conclusion: Herein, we evaluated the predictive capability of peripheral blood-based DNA methylation models for radiographic OA progression in the JoCoOA. Future work should focus on evaluating differential DNA methylation of individual peripheral blood immune cell populations in individuals with knee OA.
 Model performance characteristics
Model performance characteristicsDisclosures: C. Dunn, None; C. Velasco, None; L. Schlupp, None; E. Prinz, None; V. Izda, None; L. Arbeeva, None; Y. Golightly, None; A. Nelson, None; M. Jeffries, None.

